Basic Information
- Novermeber 7, 2018
- Howard Hughes Medical Insititue Investigator
- David B. Arnold Jr. Professor of Science of Chemistry and Chemical Biology
- Professor of Physics
- Harvard University
Abstract
通过成像在纳米尺度和系统尺度上启发(照亮)生物学
作为生命的基本单位,细胞由许多不同类型的分子组成,形成复杂的相互作用网络。因此,解剖细胞的内部工作需要成像方法,这样细胞内的分子相互作用就可以直接可视化。这是一系列独特的挑战。在本演讲中,庄小威教授将介绍她们为克服这些挑战而开发的两种成像方法以及这些方法的生物学应用。首先介绍STORM,一种克服衍射极限的超分辨率成像方法。然后,她将描述MERFISH,一种单细胞转录组和基因组成像方法,它能够绘制细胞内转录组和基因组的空间组织,以及复杂组织中不同类型细胞的组织。
Introduction
- Scale:
- Human -> Cells -> Molecules
- 1 meter -> 10-100 microns -> 1-10 nanometers
- Target:
- want to visualize the interaction of these molecules
- want to figure out how do they form intricate networks that give life collectively to a cell
- Post the problem:
- If you want to do it with a direct visualization approach
- and want to think about how difficult that task might be
- Give an analogy:
- enlarge the human size to the whole Earth
- the size of a cell is like a district equivalently of the Berkeley campus
- the size of a molecule is like a head of a human
- the action of peeking inside living systems to see these molecules is as if you are an astronomer looking into a distant plantet with a very powerful telescope
- and you not only need to see that planet but you want to see the head of whatever life form is on that planet
- Desired properties of the ideal visualization methods
- Molecular-scale resolution
- beacuse molecules are really small (mentioned in the analogy)
- Molecular specificity
- there various types of mulecules inside the cell
- need to know the identity of these molecules
- Dynamic imaging
- we are dealing with living systems, which are changing and moving all the time
- we need to catch these dynamics
- Molecular-scale resolution
- After considering these three requirements, it is natural to think about light microscopy
- for dynamic imaging
- light is relatively gentle and can imaging without perturbing the living systems
- and it also has reasonably fast time resolution so you could catch these processes in real time
- for molecular specificity
- light is wonderful for it has so many different colors
- there are many different colored fluorescent probes that can be linked to your biological molecule of interest with high specificity
- for dynamic imaging
- Post the problem that it used to be: light microscopy did not reach the molecular scale resolution
- it is because of a well-known phenomenon to physicists, that’s diffraction
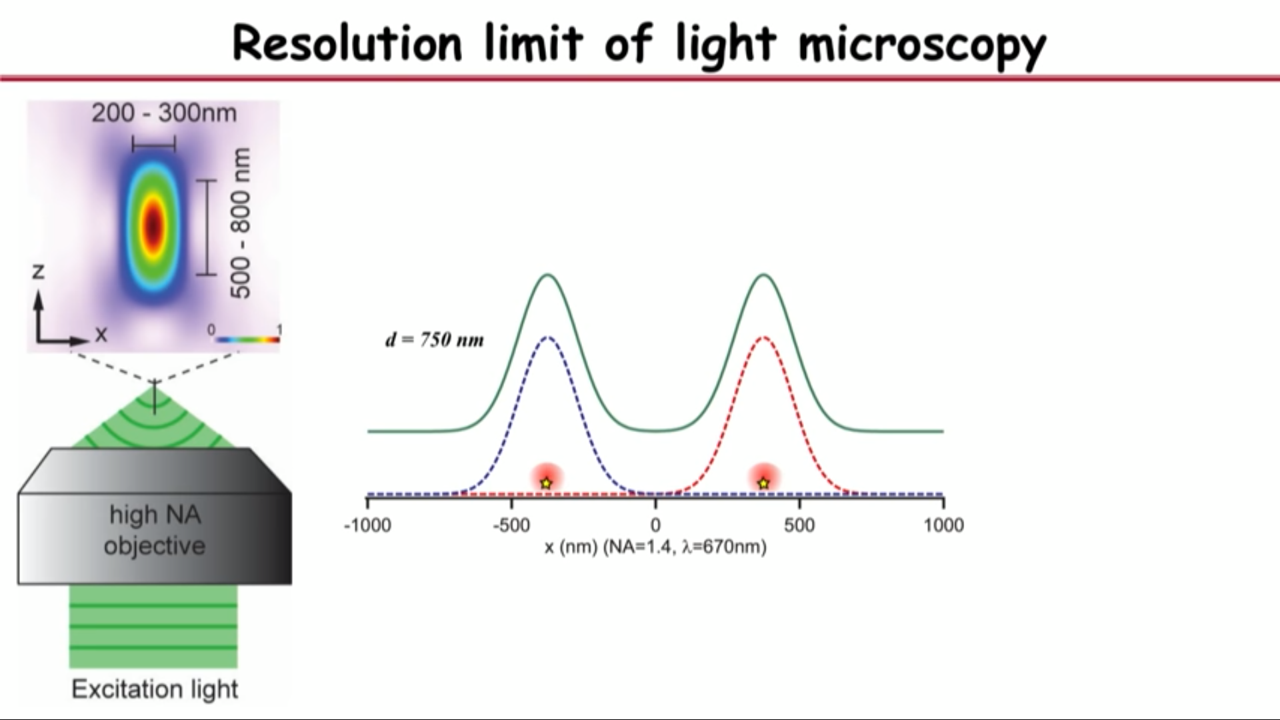 | 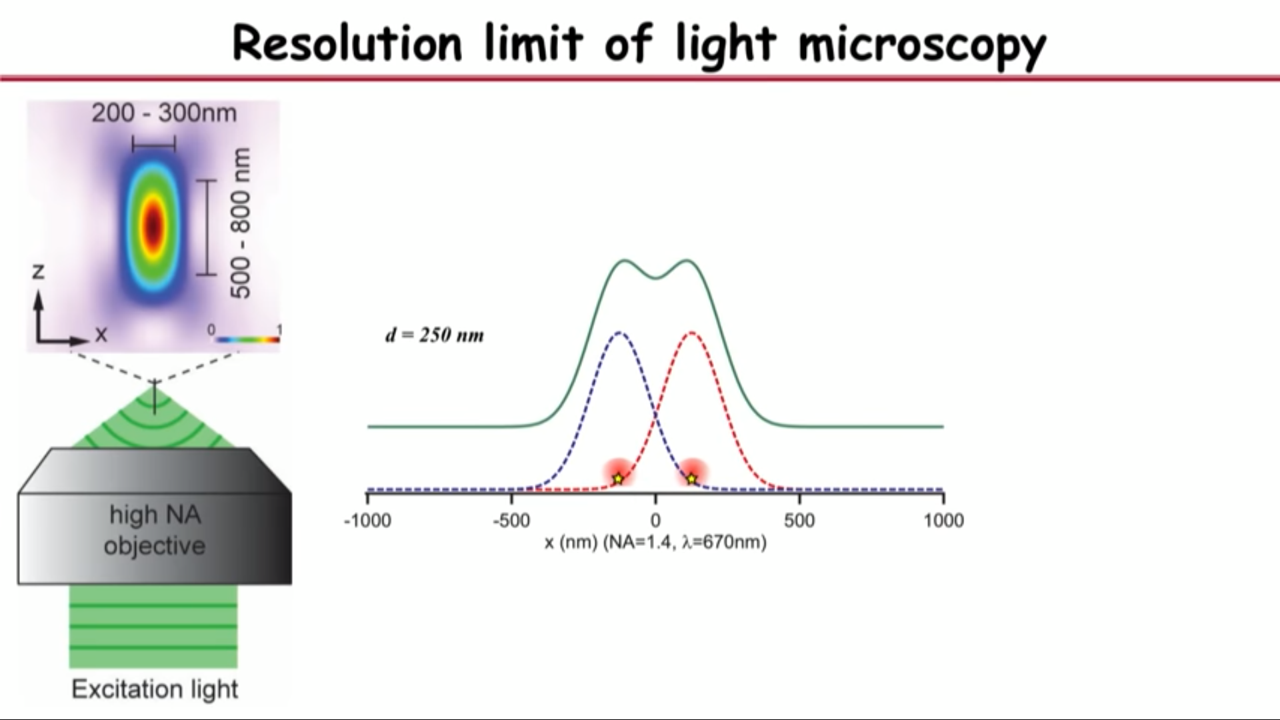 | 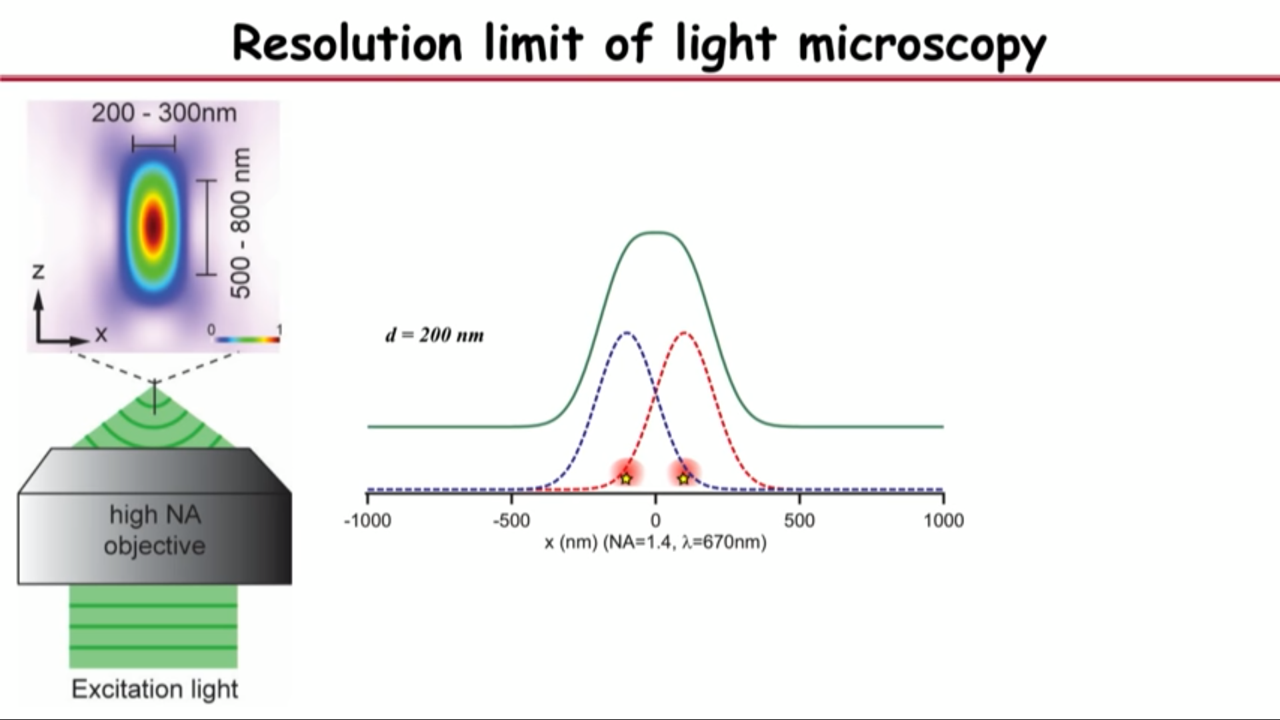 |
- Resolution limit of light microscopy
- no matter how small an object you are imaging, be it even a single molecule, its image will have a finite size of about a few hundred nanometers
- if you have two objects that are far apart, their images will not overlap and you can resolve them
- but as you get them closer, eventually their images will merge into a single blob and that’s when you are no longer capable or resolving it
- Recognized in the 1870s by Ernst Abbe: this diffrection limit is also called Abbe resolution limit
- Introduce to Super-resolution imaging
- this resolution limit has been overcome by multiple type of methods (Optically there are two major categories of approaches)
- one is using spatially coordinated manner to separate molecular signals: Patterm-illumination-based approaches (SIM)
- The other is Single-molecule-based approach
- this resolution limit has been overcome by multiple type of methods (Optically there are two major categories of approaches)
STochastic Optical Reconstruction Microscopy (STORM)
belongs to Single-molecule-based approach
Even though the image of a single molecule is diffraction limited, you can pinpoint the center position of this image with very high precision.
Basis
- Gelles et al, Nature(1988)
- Moerner et al, PRL(1989)
- Yildiz et al., Science (2003)
But it does not overcome the diffraction limit. Because the diffraction limit. And that where Xiaowei Zhuang came in (solve the overlapping problem).
Innovation in Super resolution imaging: Overcome the diffraction limiting
- Rust et al, Nat Methods (2006) - STORM
- Betzig et al, Science (2006) - PALM
- Hess et al, Biophys. J (2006) - FPALM
Take the advantage of the other dimension.
Molecules’ image could overlap in space. But if we take advantage of the time dimension, then we can separate them in time so that in both space and time, they do not overlap.
So what Xiaowei Zhuang’s group do is
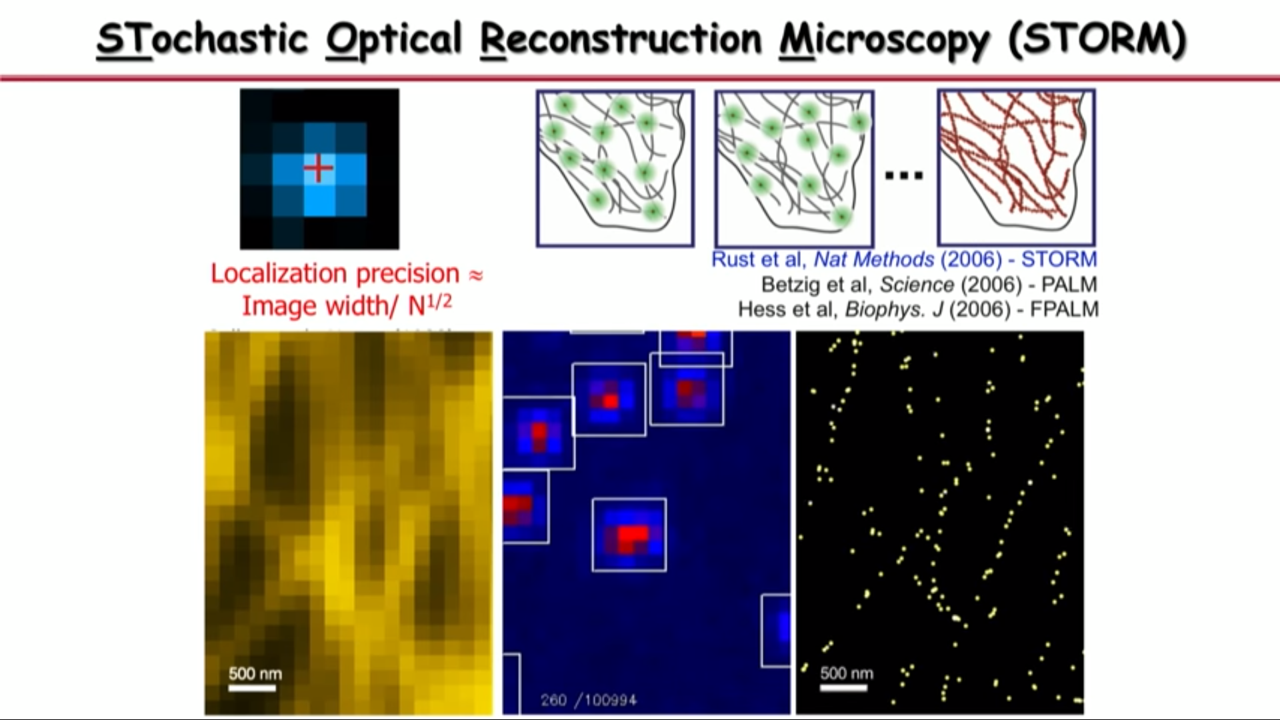 | 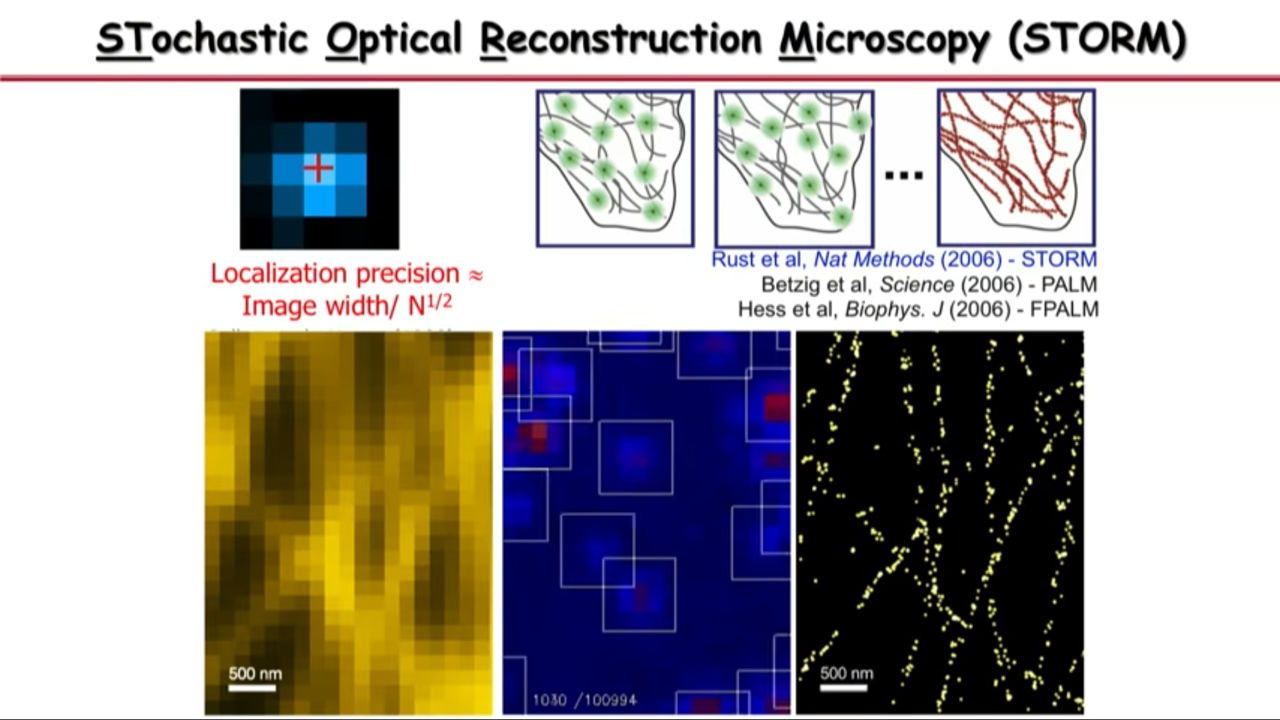 | 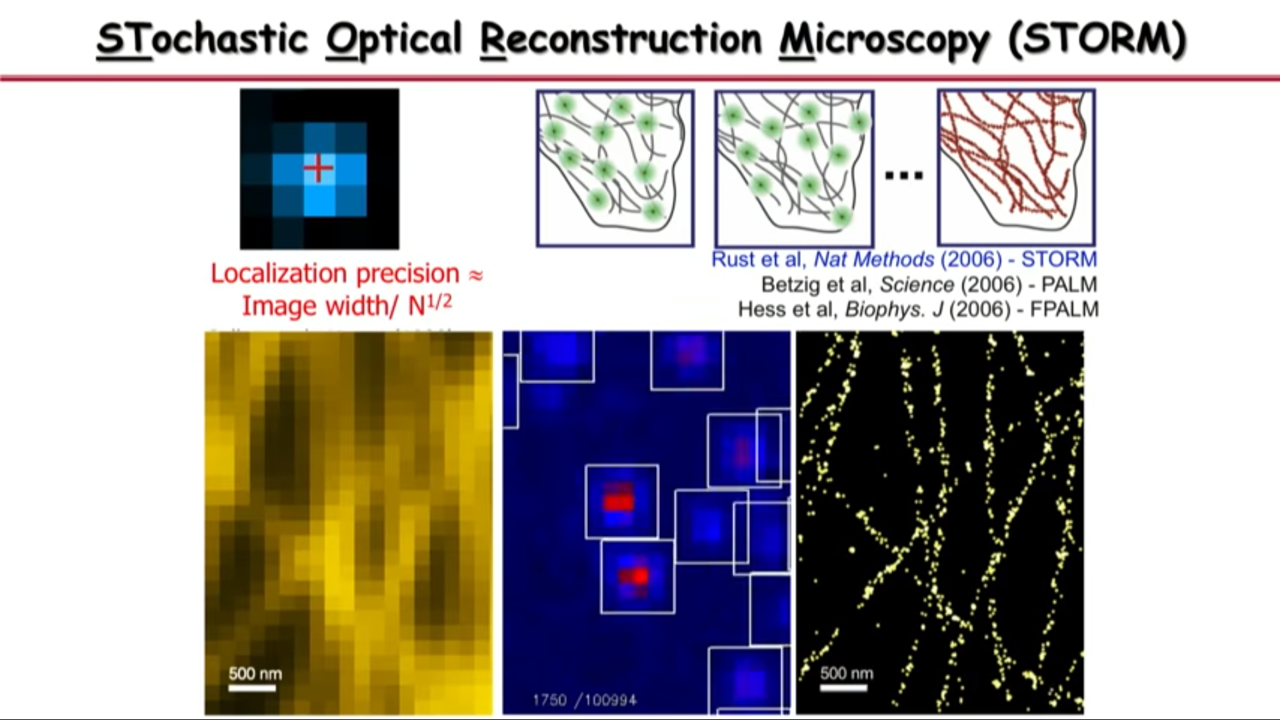 |
- at any time they can activate a subset of molecules so that their images are no longer spatially overlapping, they pinpoint their center position with high precision,
- and then they turn off these molecules
- and turn on a different subset.
- And they iterate this thousands of time until their determined all the molecular positions in the field of view.
- And then with that you can see that the image is no longer limited by diffraction or by how wide these single-molecule images are, but by how precise you can localize these molecules.
What led them to this idea:
- Their group discovered that some fluorescent dye molecules are photo switchable. They have different physical conformations or different chemical state.
- In one state, if you shining light, they can fluoresce.
- In the other state which they call dark state, even if you shine light on them, they won’t fluoresce.
- But they can be activated from dark state to fluorescent state by a different wavelength of light.
- And then because they are separated by a energy barrier, if you use a weak enough light, then you can get a small fraction of them that go across this barrier into the fluorescent state.
- This is how they can achieve what they call sparse activating or activation or activation of a subset of molecules.
Details:
- They don’t control which subset (small subset, 0.1%). It is a stochastic subset. -> “Stochastic Optical reconstruction microscopy”
- Similar methods have been independently developed and published nearly simultaneously by two other teams.(PALM, FPALM)
So far I have told you ways to get a super resolution image in two dimension. But basically all biological structures are three-dimensional. We not only want to overcome the diffraction limiting in two dimensions in XY, but also want to do that in the third dimension, the Z dimension. (Xiaowei Zhuang)
Innovation in Super resolution imaging: Overcome the diffraction limiting in three dimension
3D STORM
- Huang et al, Science (2008)
- Vaughan et al, Nat Methods (2012)
- Xu et al, Nat Methods (2012)
- Jia et al, Nat Photonics (2014)
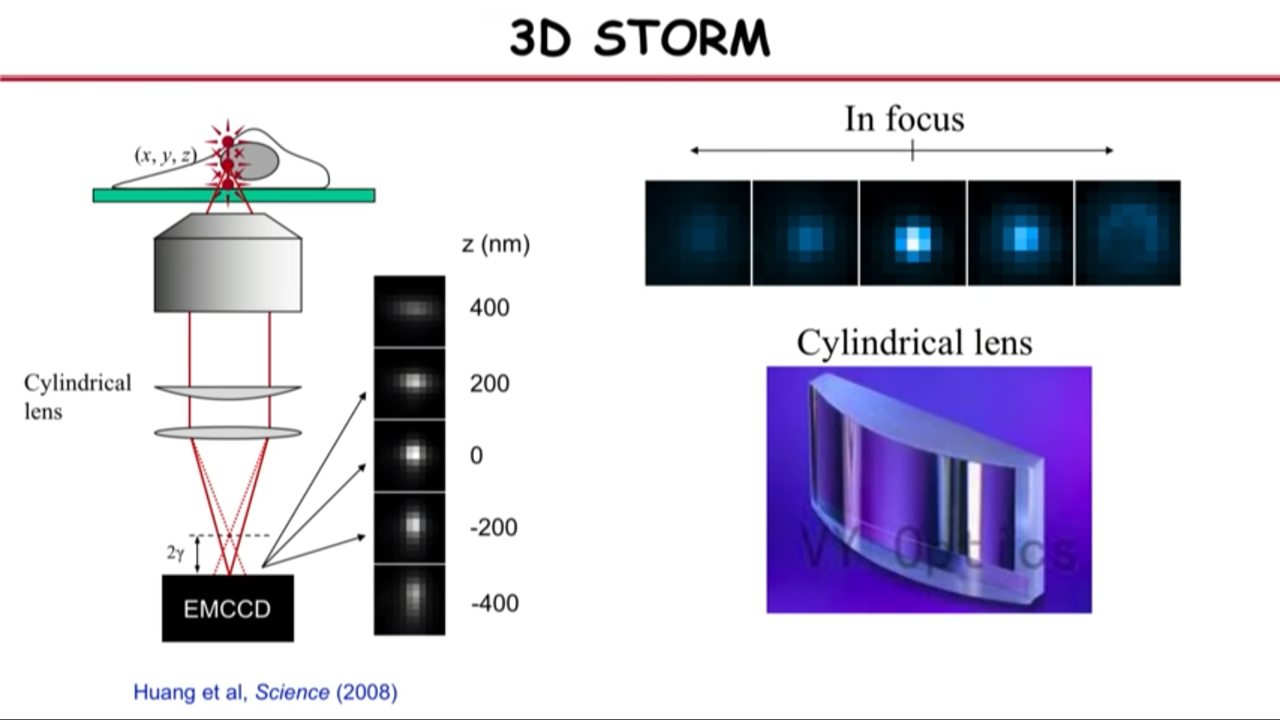 | 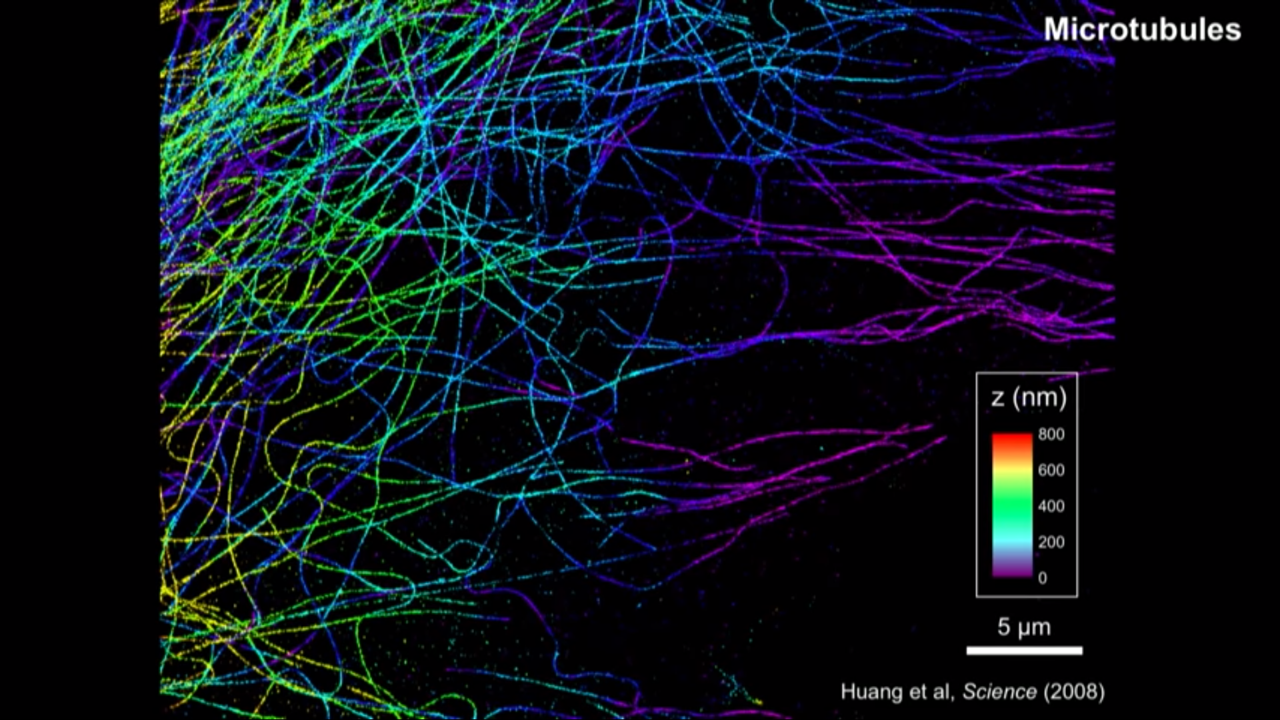 | 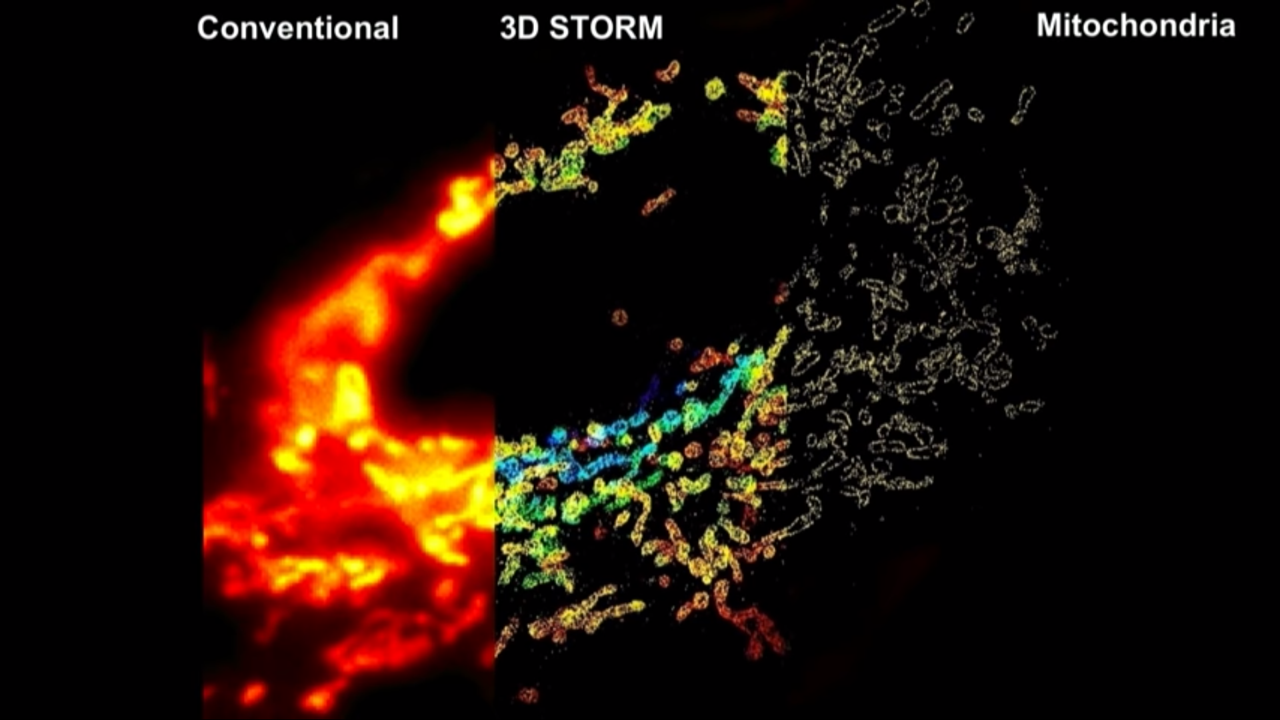 |
How to get the third dimension information:
- with lenses in and out of focus
- the images in focus are sharp
- the images out of focus are blurry
- for single-molecule images, that blurriness basically means the image looks darker and broader
- in other words, the molecules in differnet Z, the shape of their images encode their Z information.
- The problem of this is that the in focus is the local minimum and the slope is zero. And below and above focus, they look the same, so there is a degeneracy
- their group actually used a very simple approach to overcome this problem, and that is they insert a cylindrical lens into the detection path.
- The cylindrical lens only bend light in, its only curved in one dimension.
- With that they can actually turn individual molecule’s images from spherical into an ellipsoidal picture form
- That ellipticity depends very sensitively on the Z position
- With that they can actually do all three dimensions super resolution imaging.
Development:
- over the years, their group improve it with either the development of new imaging schemes or the development of different molecular probes that brighter then indeed now for some system we can get to this a few nanometer resolution, which is at the true molecular scale. And then indeed now that’s two orders of magnitude better than the diffraction limit
- there are many other labs who has been constantly doing this kinds of improvement too
- Worth mentioning: Stefan Hell’s new method called MINFLUX
- MINFLUX actually gets to one to two nanometer resolution
- Live-cell STORM
Applications: Link to biological discoveries
I am pretty firm believer of that saying that if you invent a new method, no matter how cool it is, if it dosen’t lead to scientific discovery, it’s still nonetheless a useless method.
 |  |  |
Detailed Example: Discovery cellular structures that you did not even know existed before
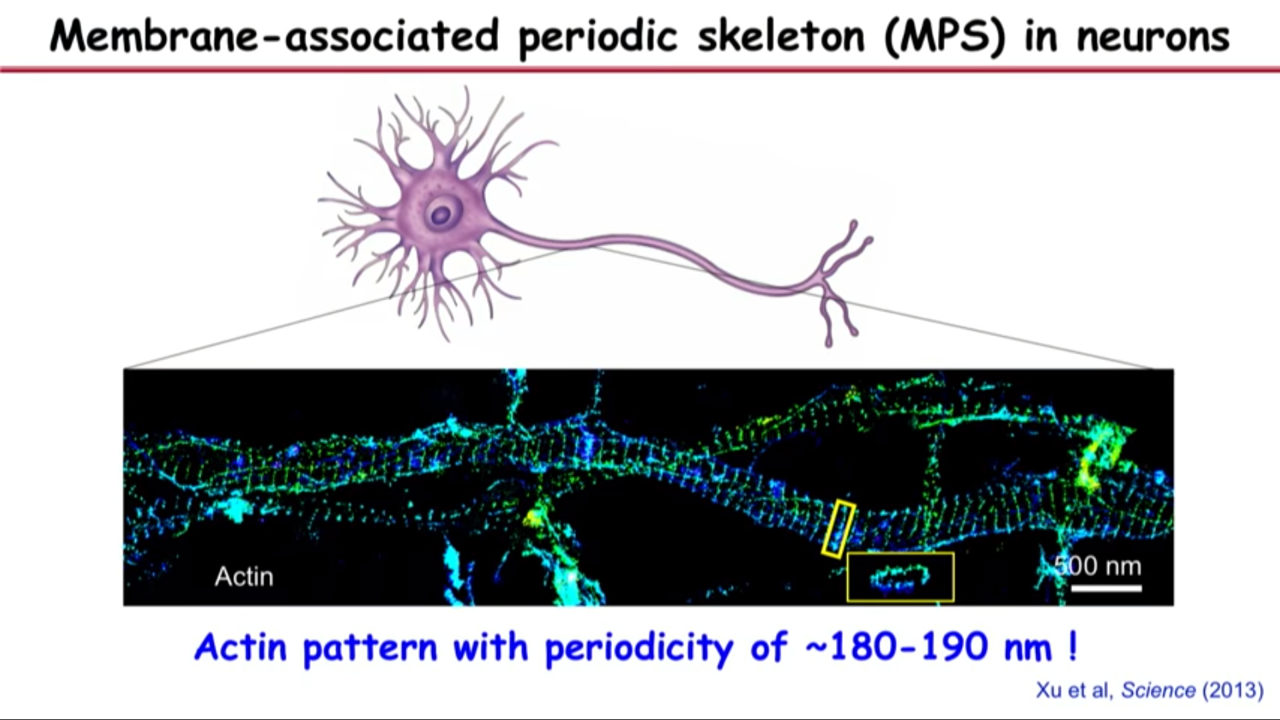
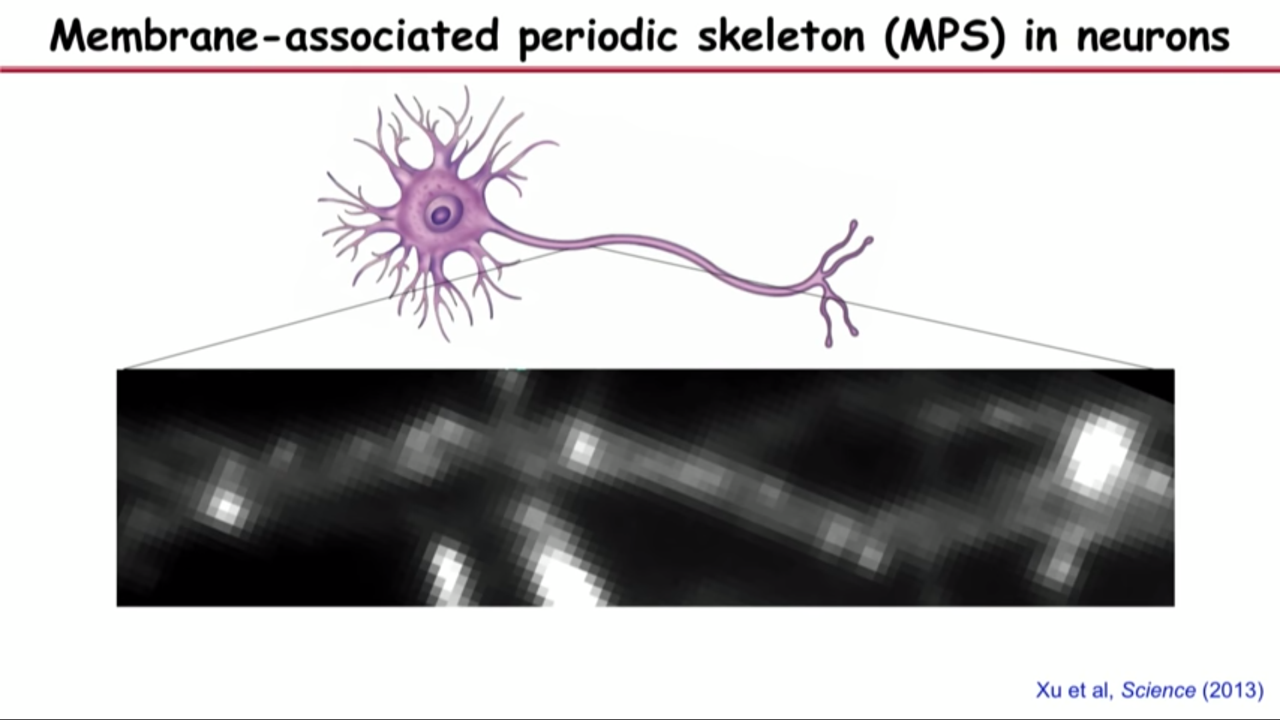
- Membrane-associated periodic skeleton (MPS) in neurons
- This band structure is the most prominent actine structure that we can see in axons.
- These bands are actually, if you turn them around, they are little rings that wrap around the circumference of the axon right under the axonal membrane.
- The reason that why you cannot see this structure before is if you do the same image in the same field of view with diffraction limit optics, you would completely obscured the structure.
- That because the periodicity of the space between these bands are just below the diffraction limit.
- 180 nanometer spacing (between bands) also gave them clue of what other molecules could be in the structure becaues if you see these actin rings periodically spaced, then you couldn’t help wondering whether there might be a molecular spacer that are connecting these actin rings.
- And one kind of molecules (if there is), the spacer has to satisfy two requirements: 1) it is 180 nanometer long; 2) it should interact with actin.
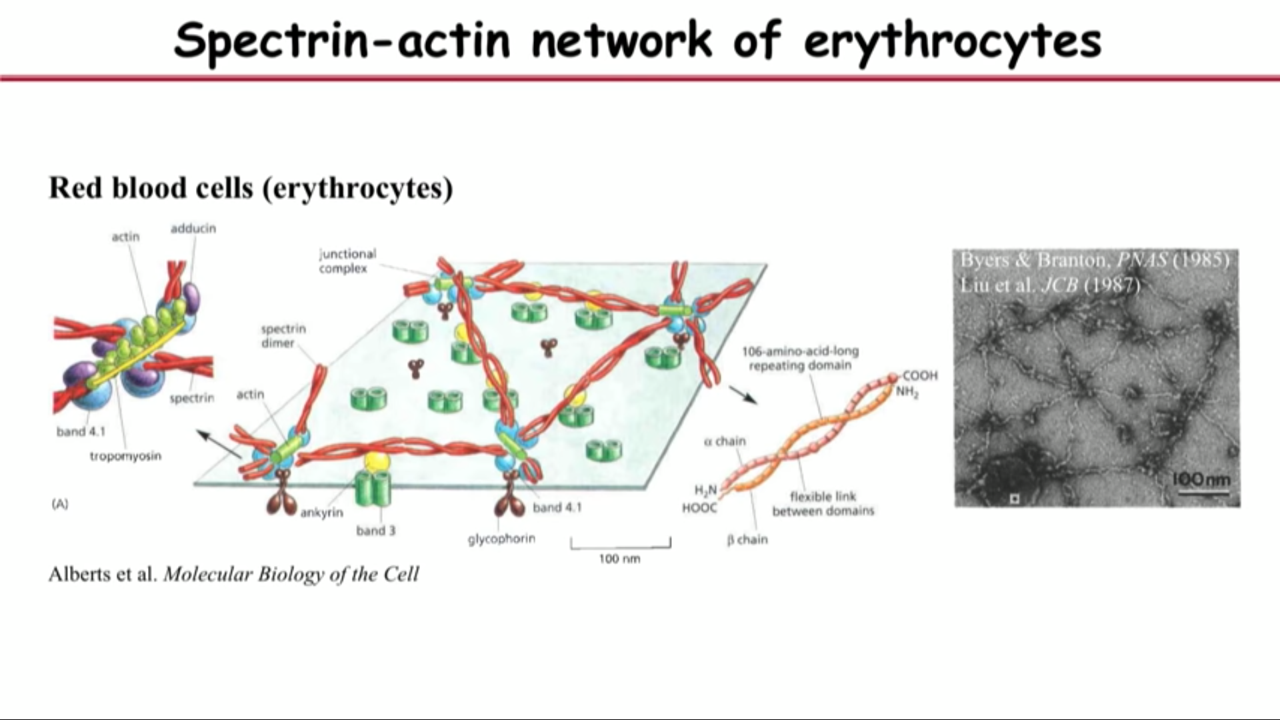
- the molecular that satisfy these two requirements is called spectrin
- the best we know about spectrin is actually in a different kind of cell, red blood cell.
- That led them to the hypothesis that what they saw are these little actin filaments aligned into ring structure. And the reason they are periodically spaced is because they are connected by the spectrin tetramers.
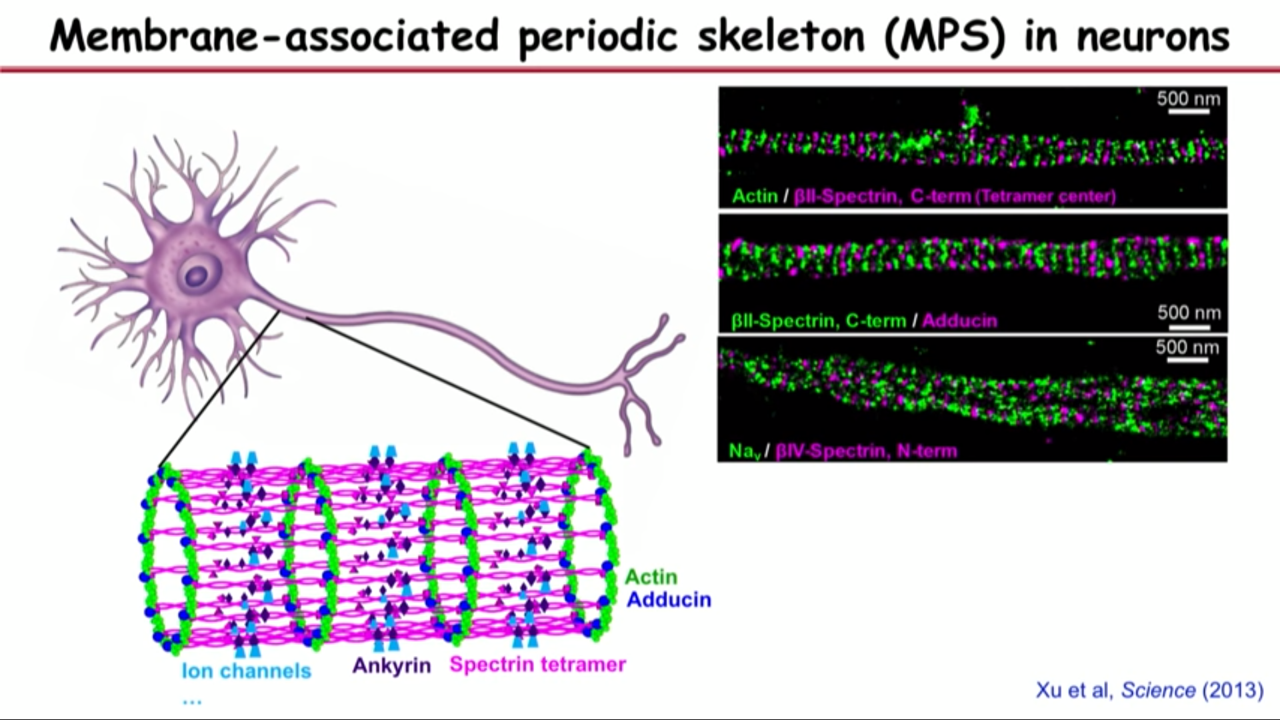
- A good way to test the hypothesis is with multicolor STORM imaging where they imaging both actin and spectrin.
- They can not only imaging spectrin but also imaging the middle portion of spectrin which they could recognize was an antibody, then they would expect alternating green, magenta ring. (-> Alternative green magenta bands)
- And later found many other molecules on the structure
 |  |  |
Prevalence of the MPS
He et al, PNAS (2016)
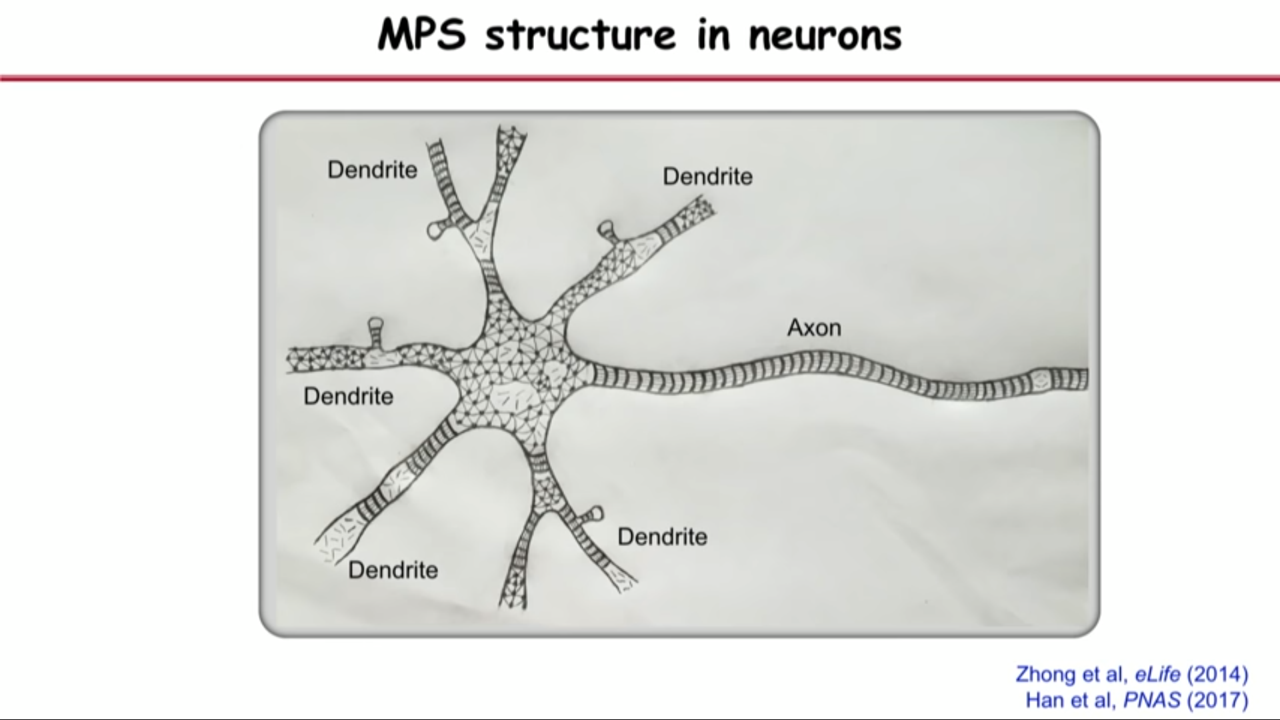
- The MPS is present in many different types of neurons
- excitatory and inhibitory neurons
- central and peripheral neurons
- myelinated and unmyelinated neurons
- The MPS is conserved in diverse animal species
- C elegans
- Drosophila
- Rodents
- Human
Functional roles of the MPS
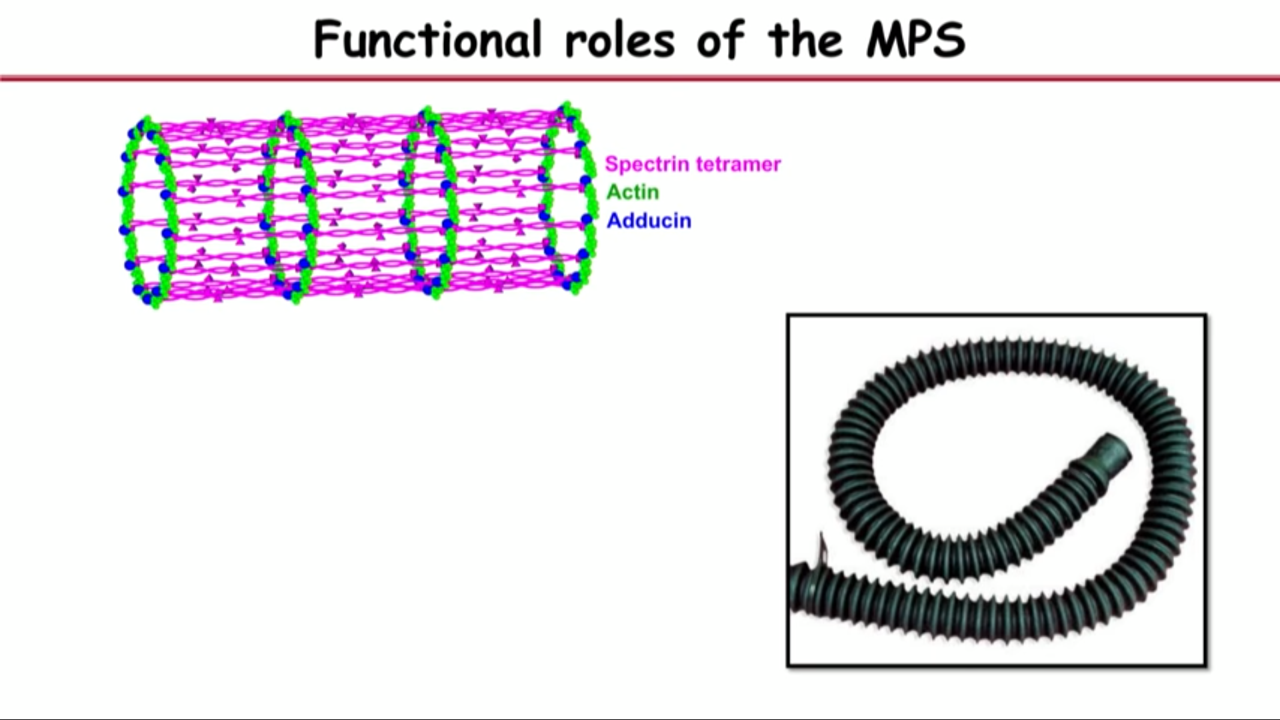 |  | 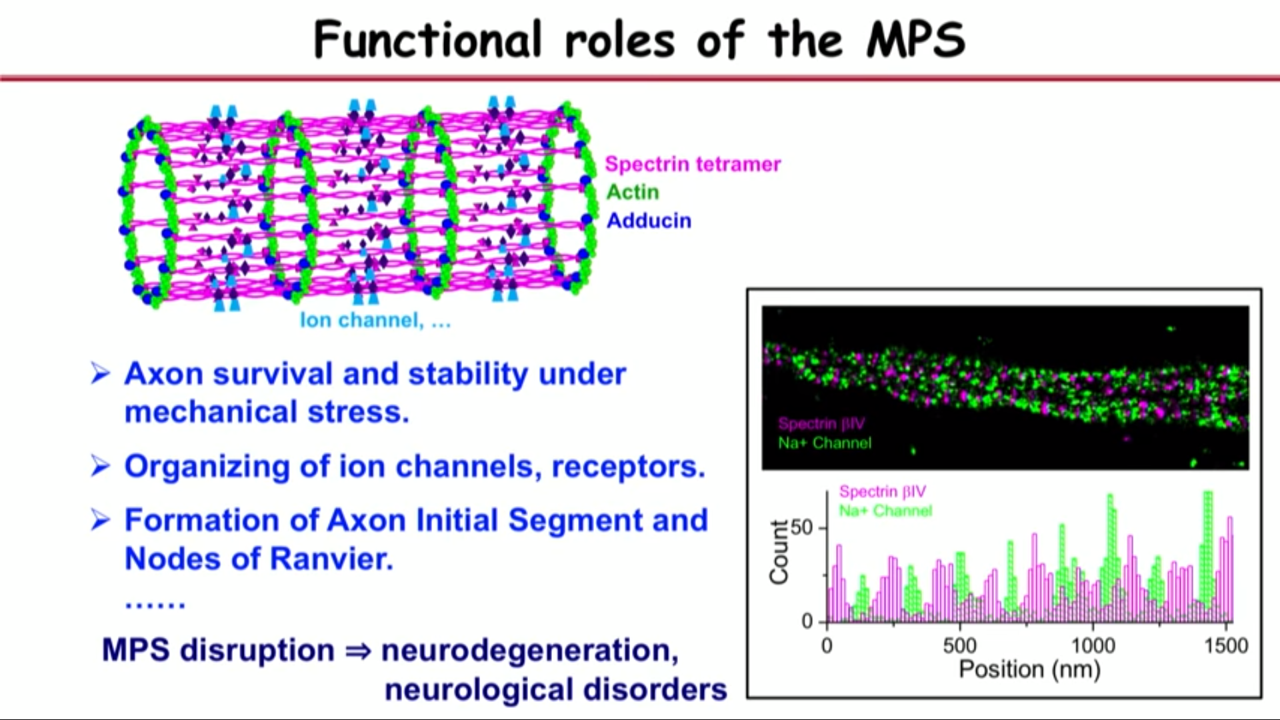 |
analogy to vacuum hose
- If you have a rigid ring connected by a flexible material, then you can bend and stretch it without breaking it
- This might be indeed what a structure like axon or dendrites needs.
- Because these can be really thin structures and very long so it can be brittle.
- And they can be constantly subject to mechanical stress
- Need a flexible yet durable skeletal structure like this to support the axons.
- Experimental data pointing towards that:
- if you delete this key molecule spectrin in a little animal called C elegans and then the axons actually tend to spontaneously break
- Interestly, if you actually make these animals paralyzed so that they cannot move, but they can develop into full shape, the axons can actually grow into proper morphology, suggesting that the structure can give the axon its stability under mechanial stress.
- …
Development:
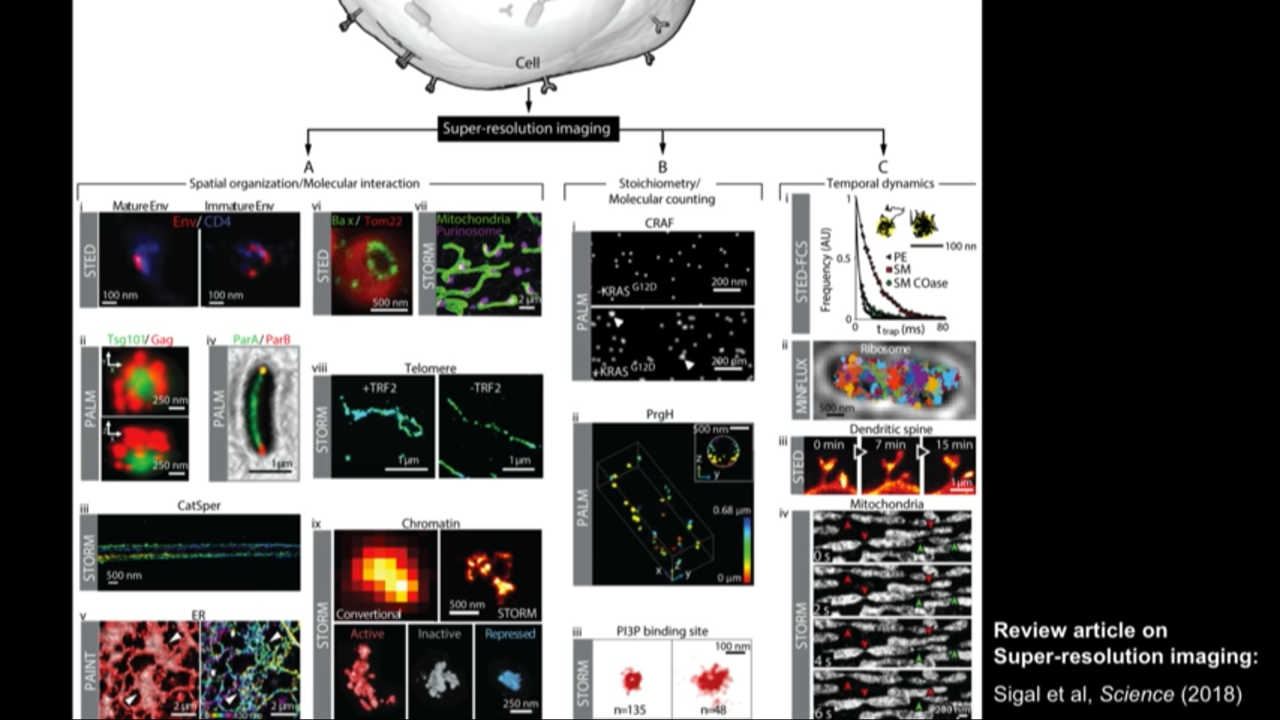
Super resolution field is a big field rapidly growing now.
Review article on Super-resolution imaging: Sigal et al, Scinece (2018)
 |  |
Molecular architecture of the cell
We hope that some day, we will have a comprehensive picture of the molecular architecture inside the cell unraveling in front of us just like how we watch movies of these things.
And of course with that, we would need the molecular and sub-molecular scale resolution that needs to be routinely obtained in biological systems and to directly reveal molecualr interactions.
Also need high time resolution and long imaging time tominitor complete molecular processes in real time
One thing that I want to emphasize again is inside our cells, there is not just one kind of molecule or two or three kinds of molecules but actually various kinds of molecules. It would be great if we could see these molecules all simulataneously. In other words, we would like to have this genomic scale to visualize collective actions of all molecules and see how they give rise to this emergent property of life
They made some progress along the genomic scale imaging direction.
Genomic-scale imaging
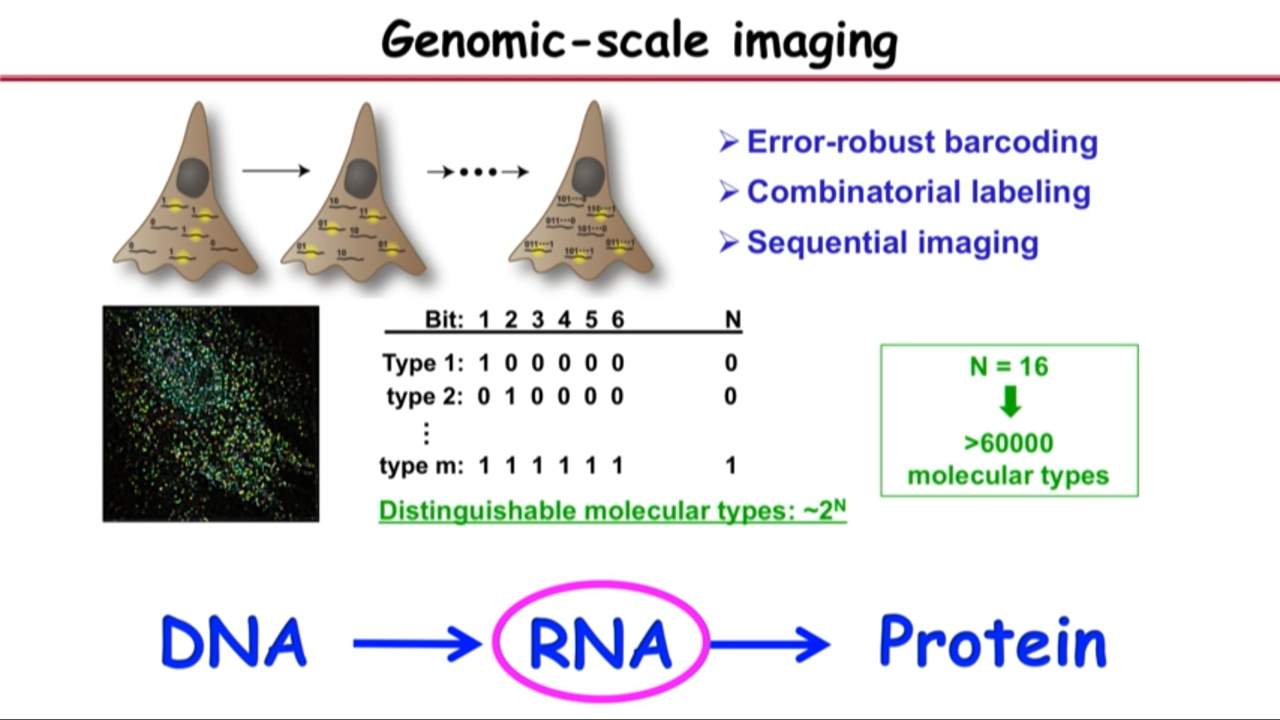 |  |  |
Concept of how to do it:
- genomic-scale means being able to image (tens of) thousands of different kinds of molecules
- But traditional fluorescence microscopy imaging cannot have this many different colors technically.
- there are so many different colors but the probes all have their spectrum to be wide enough that you cannot discern so many different color probes.
- To overcome the problem, came up with following ideas:
- Error-robust barcoding
- Combinatorial labeling
- Sequential imaging
Procedures:
- Encode these genes with unique binary barcodes.
- Each gene have a different barcode.
- Imprint this barcode onto those genes. Then read out this barcode sequentially.
- In the first readout (first round of imaging), they label and image only those genes their first bit reads one (not zero).
- Then they turn off the signal and then in the second round, they read out those genes their second bit reads one but not zero.
- An so on, and then so forth.
- At the end of the imaging process, you will see the molecules in the cell all look like little dots. And each dot will have a binary barcode associated with it.
- After N-bit imaging (N rounds of imaging), this method can distinguish $2^{N}$ types of molecules.
- Just 16-bit imaging can one image over 60,000 molecular species. And that is sufficient to cover the genomic scale.
Application: Applied in the central dogma of biology (RNA).
- Biological reason: tells you what kinds of genes are expressed in ourselves (a quantitative way to identifying cell types)
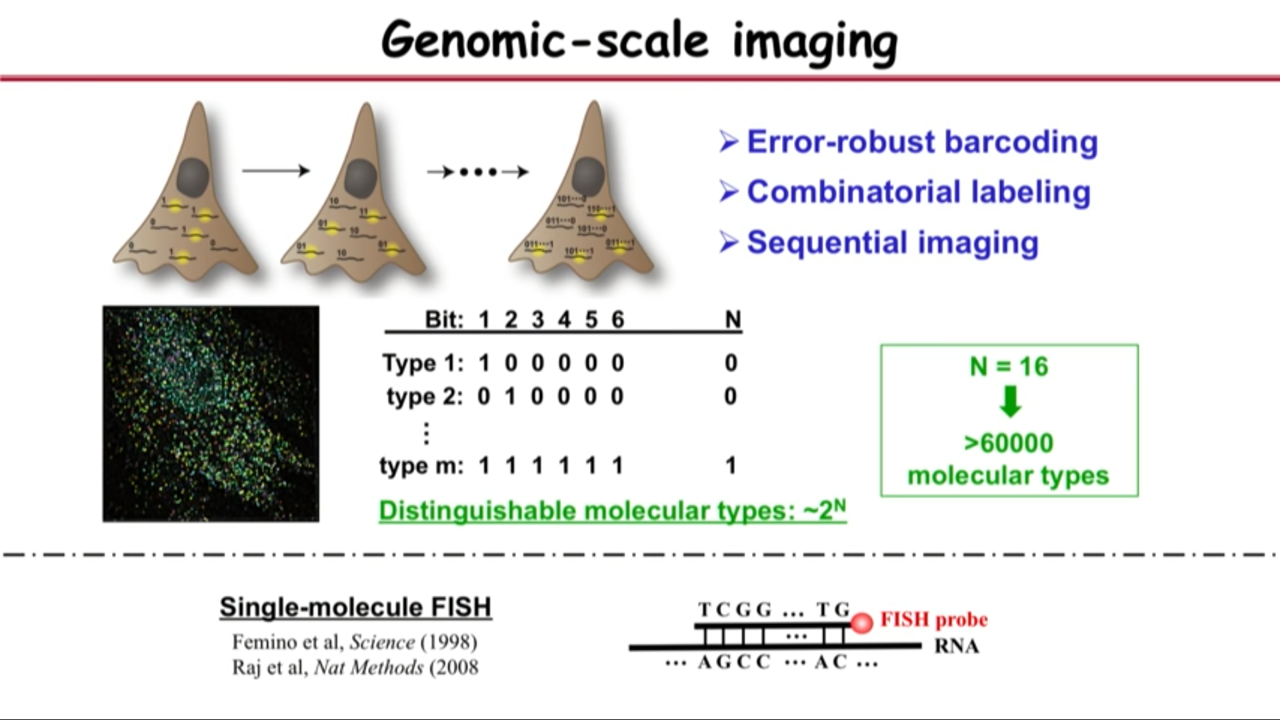
- Technical reason:
- A powerful approach for the labeling: Single-molecule FISH
- Which allows you to identify individual RNA molecules of a species can count their number inside the cell
- the process relies on hybridization
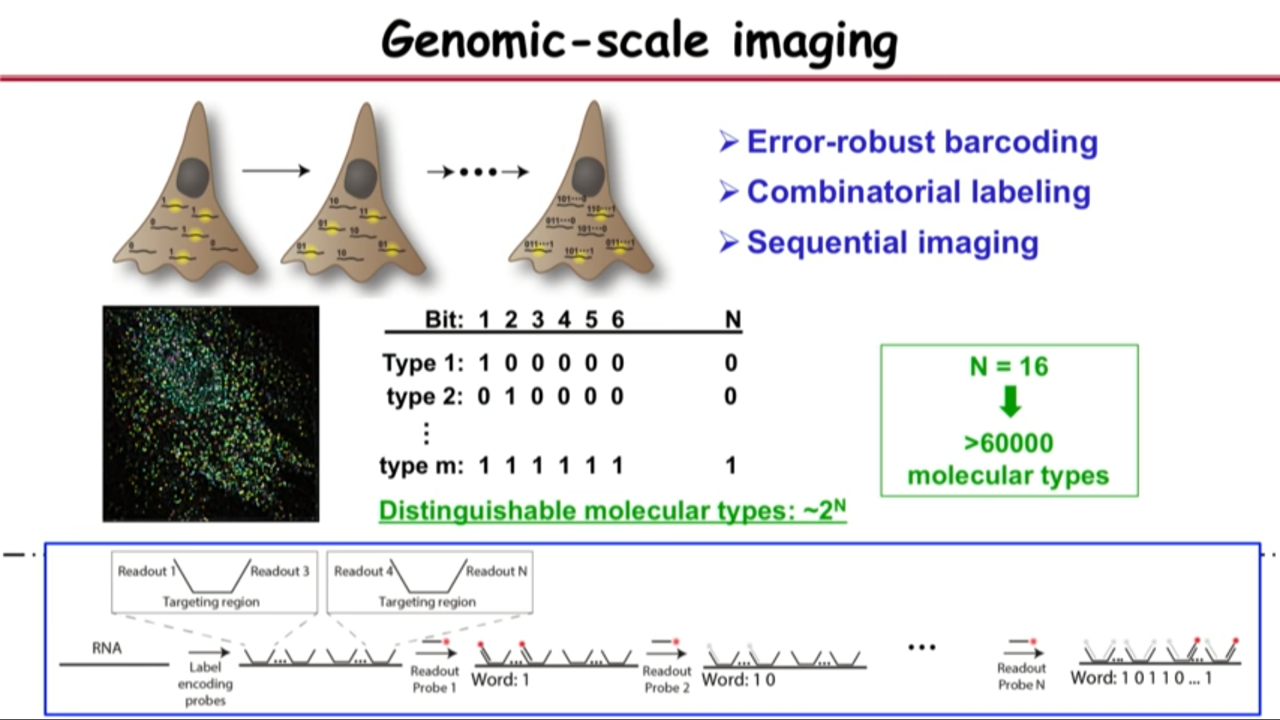
- Procedure details:
- use FISH probe to identify RNA molecules by hybridization
- these probes all have readout sequences sticking out depends on the barcode, meaning that if the barcode reads one zero one zero, it will have readout sequence one but not two, readout sequence three but not four, and so on. Then use hhybridization to recognize these readout sequence sequentially.
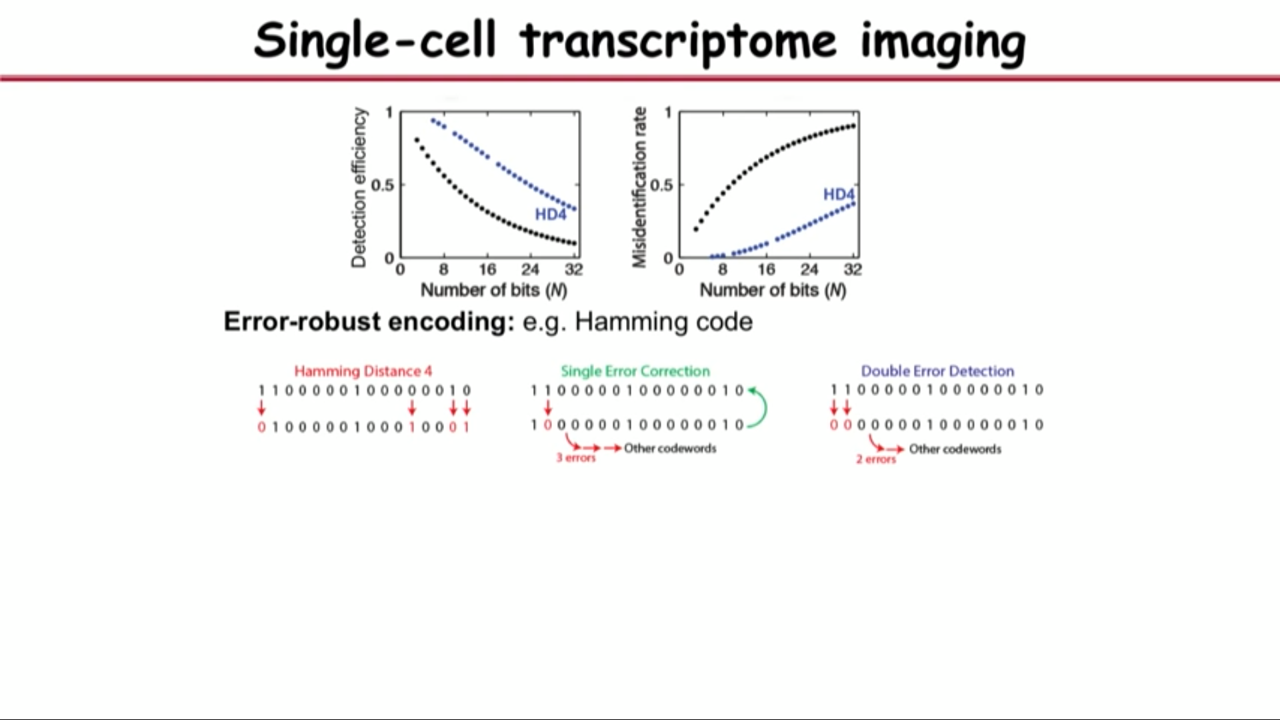
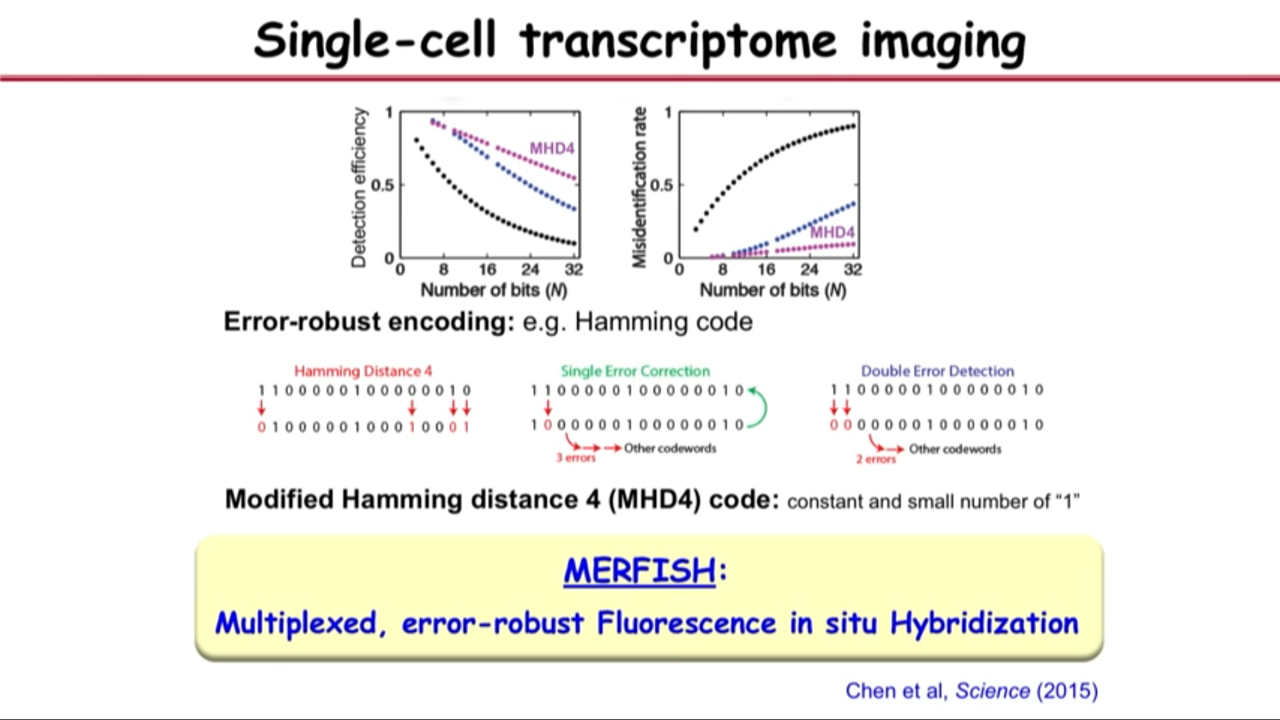
- Problem in the middle:
- single-molecule FISH is a very accurate approach (95% or better accuracy, 5% error)
- but if one bit has 5% error, two bits will have two times 5% error, 16 bits face a severe situation.
- Overcome the problem above by borrowing a trick from the digital communications.
- their prevent the erroe by using error-robust encoding
- In particular, their group implement Hamming Code
- with modified hamming distance 4 (MHD4) code: constant and small number of “1”
- and this is Multiplexed, error-robust Fluorescence in situ Hybridization (MERFISH)
 |  | 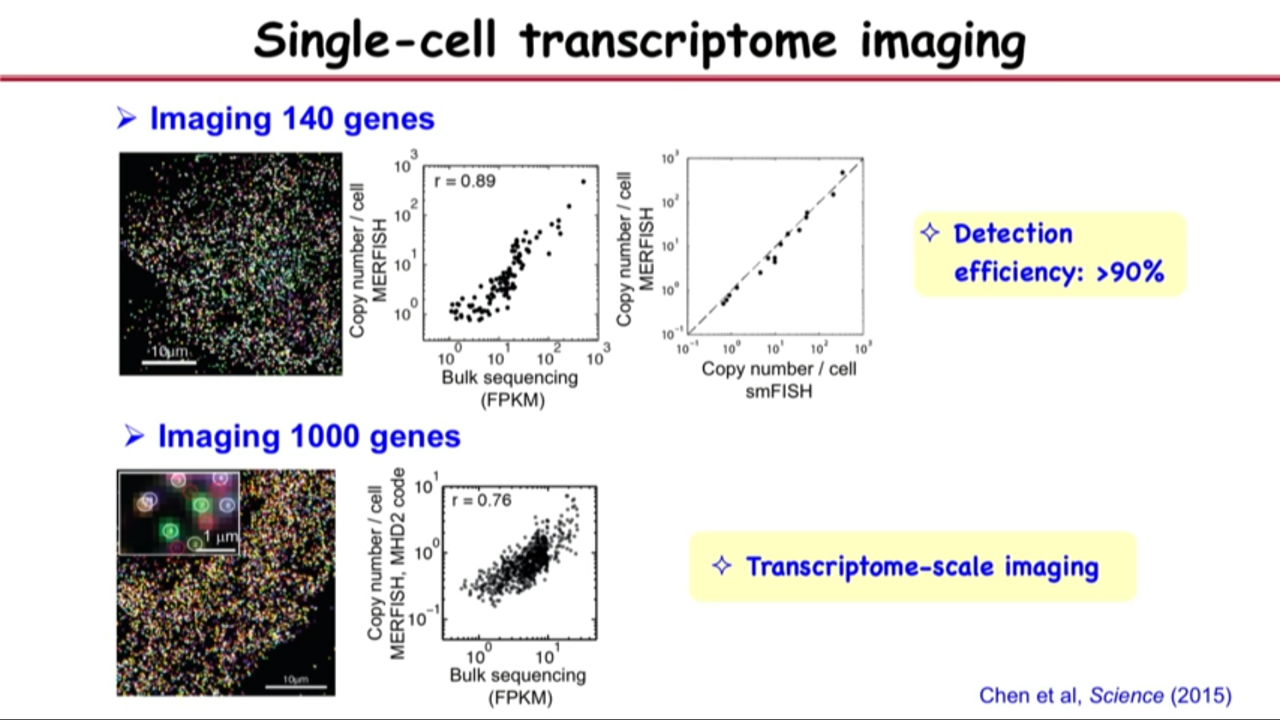 |
- Demonstrate the method (MERFISH) experimentally
- use it to image 100 genes (1000 genes or more reach the transcriptome scale) and get a high accuracy
- Detection efficiency : >90%
- use it to image 100 genes (1000 genes or more reach the transcriptome scale) and get a high accuracy
 |  |
 |  |
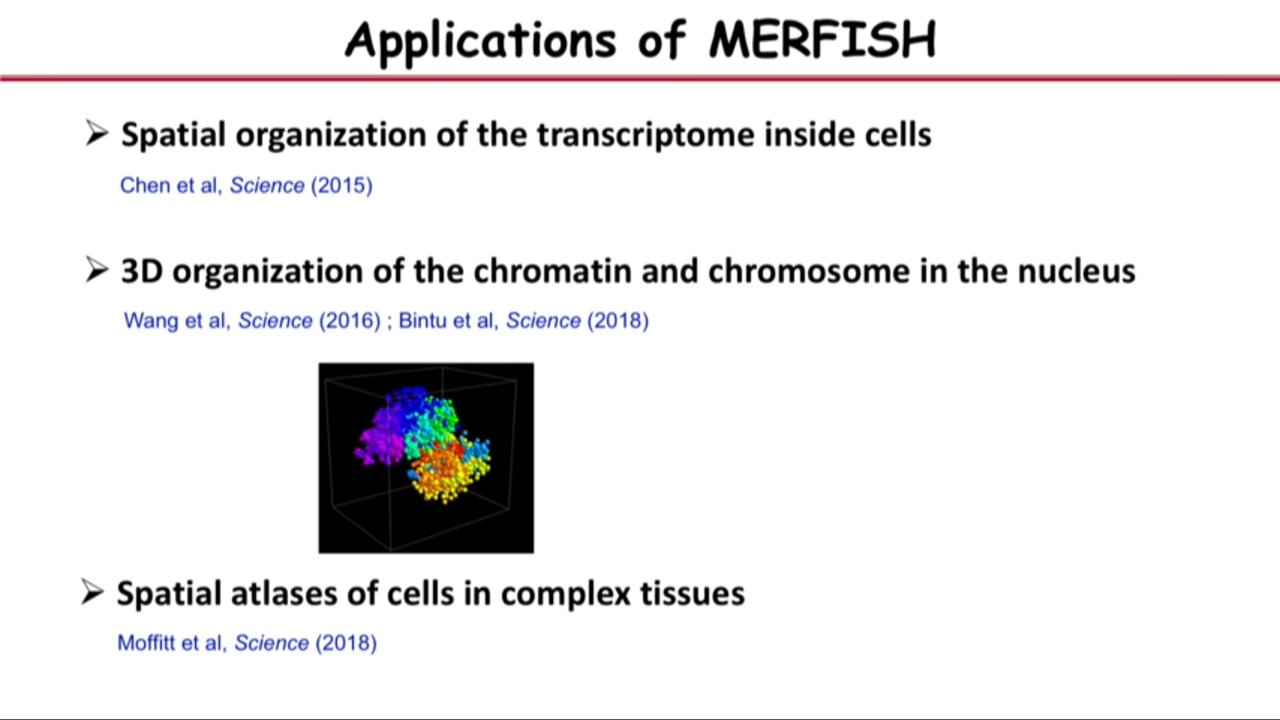
- Application of MERFISH
- Spatial organization of the transcriptome inside cells
- 3D organization of the chromatin and chromosome in the nucleus
- Spatial atlases of cells in complex tissues
- spatial organization is really key to tissue function
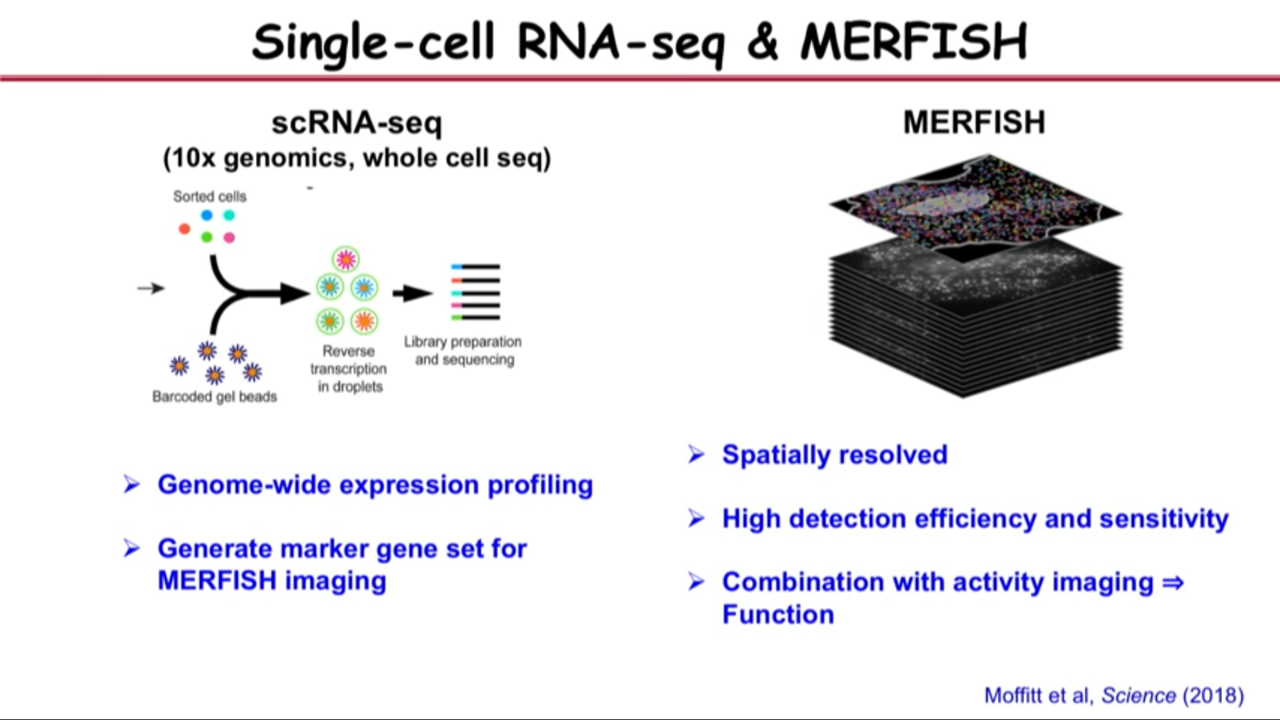
- although single-cell RNA Sequencing allow one to profile the different kinds of RNAs, their quantities in cell, and allow one to identify different types of cells, its requirement (dissociate the cell from the tissue, extract the RNA out of the cell, then subject it to a sequencing experiment) make it lost the very important spatial organization information
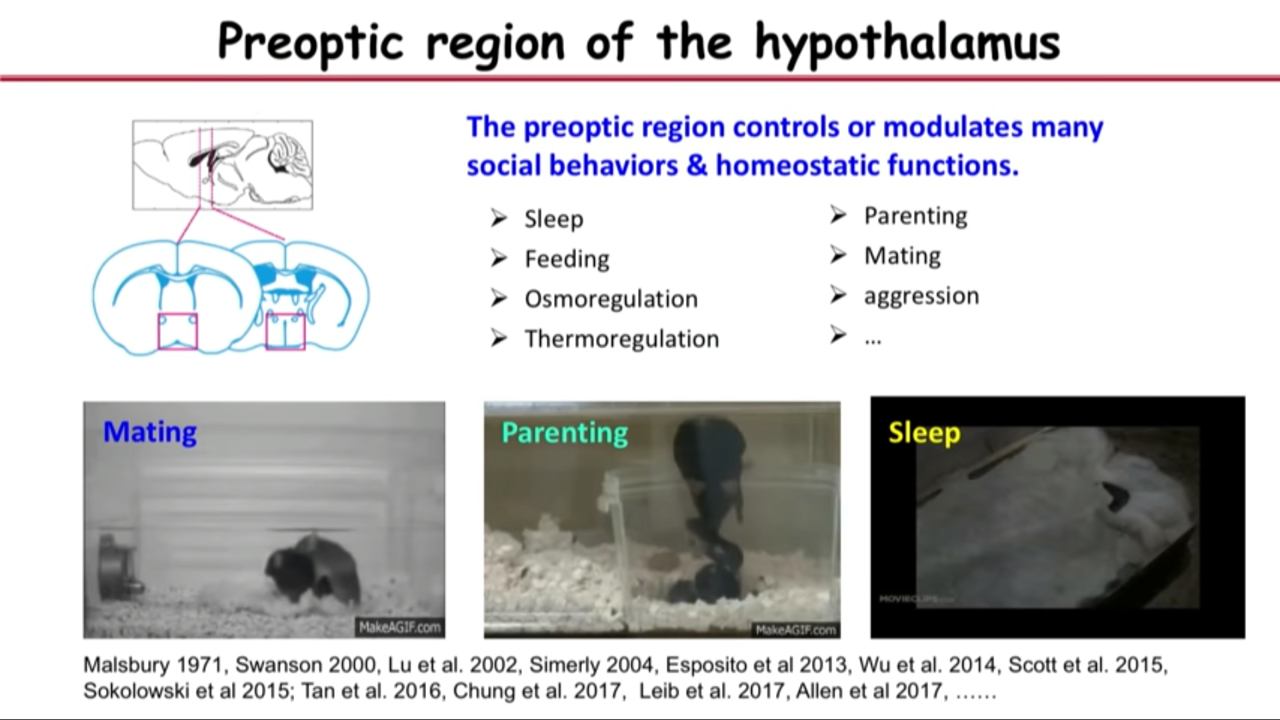
- How MERFISH can help on that front: applied to a particular brain region called the preoptic region in hypothalamus
- Combine both single-cell RNA seq and MERFISH in this study because they realized complementarities between these approaches
- Single-cell RNA seq give us genome-wide expression profiling. It help us to select the market gene sets for MERFISH imaging
- They selected the marker gene set based on both their biological knowledge of this region and single-cell RNA seq, abd then they used MERFISH to do transcriptome profiling, in situ identification of the cell types that give us spatially resolved information.
- And because MERFISH has high detection efficiency and sensitivity compared to single-cell RNA Seq, some of the genes that are expressed at very low levels that actually difficult to get by single-cell RNA Seq can be get by MERFISH. These genes are actually important for cell type, both identification and function.
 |  |  |
 |  |  |
- Application of MERFISH
- Spatial atlases of cells in complex tissues
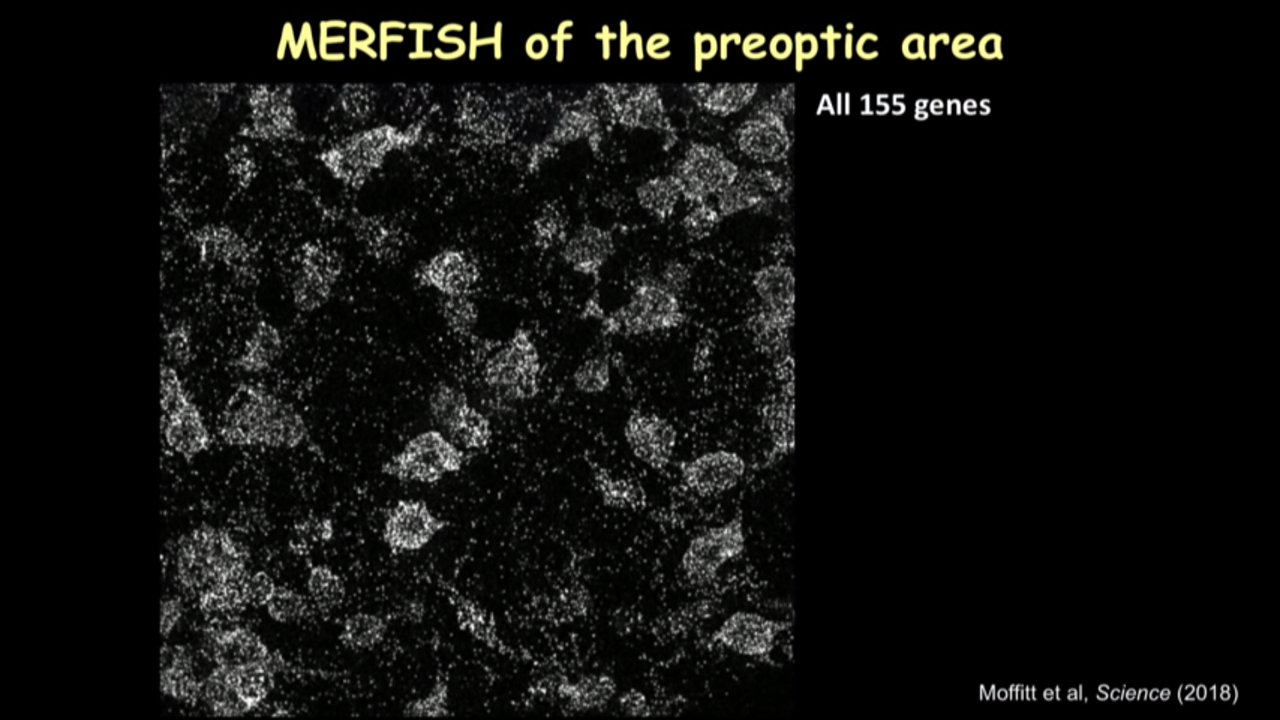
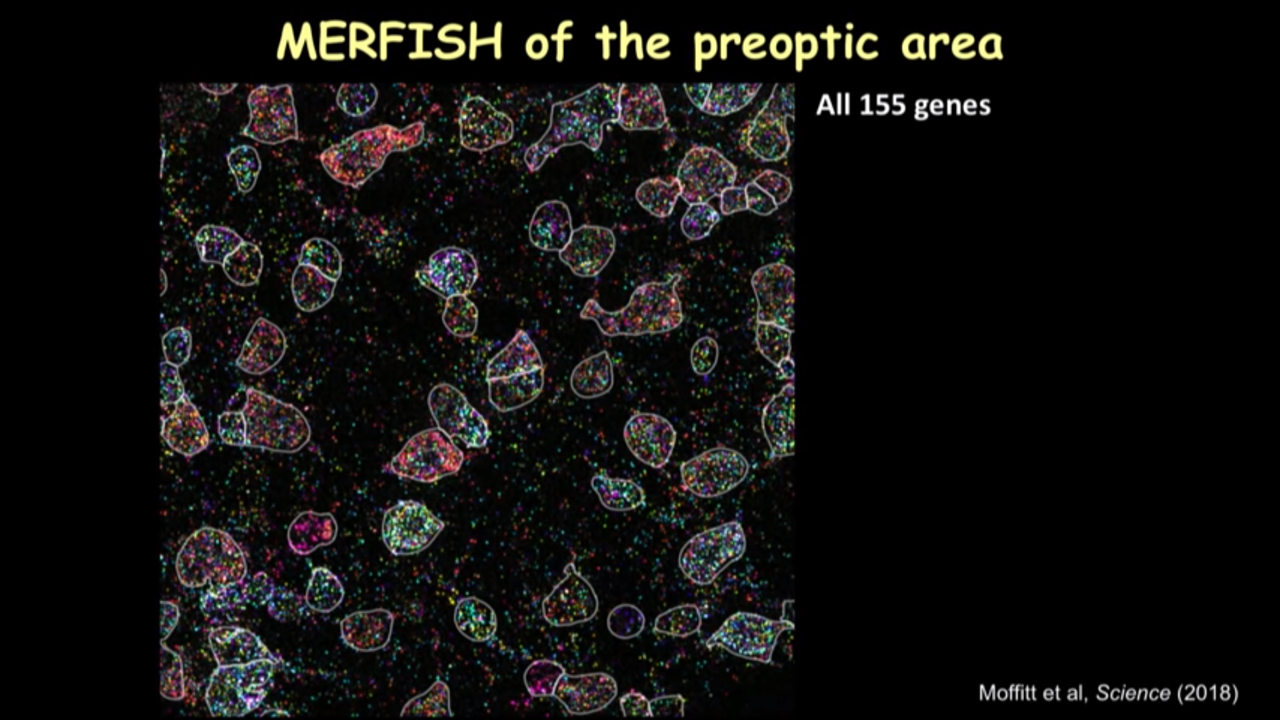
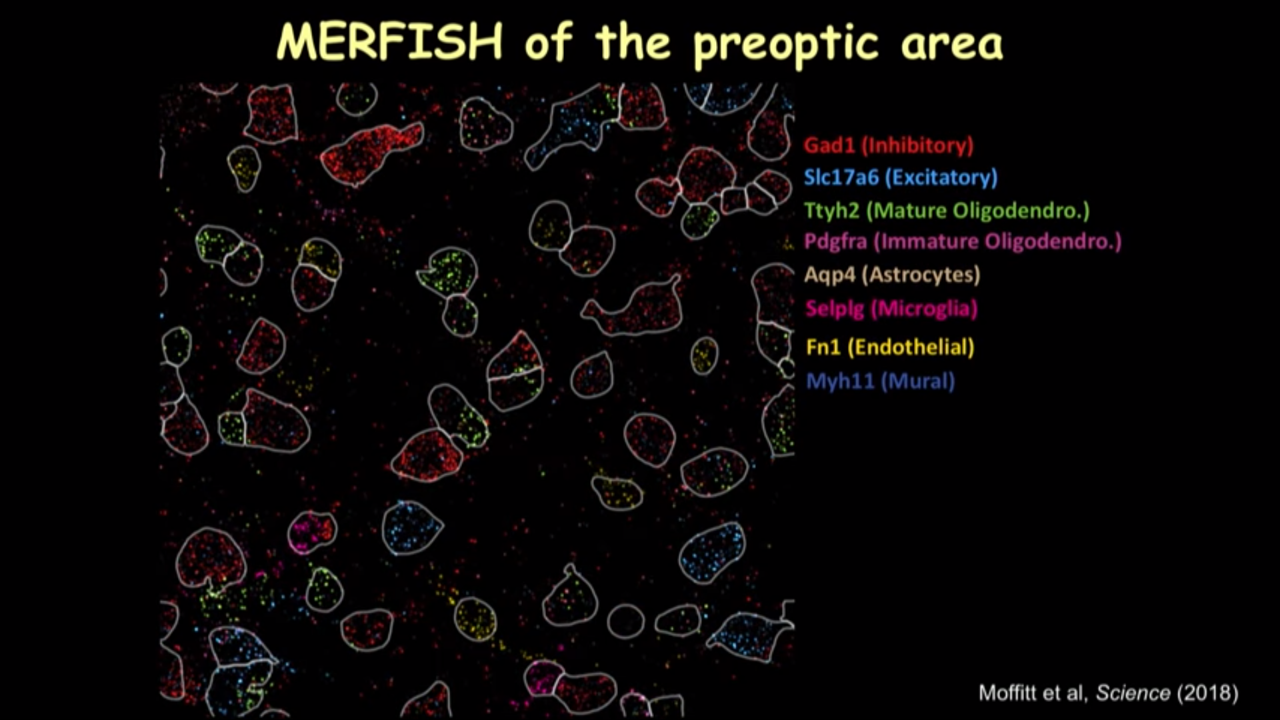
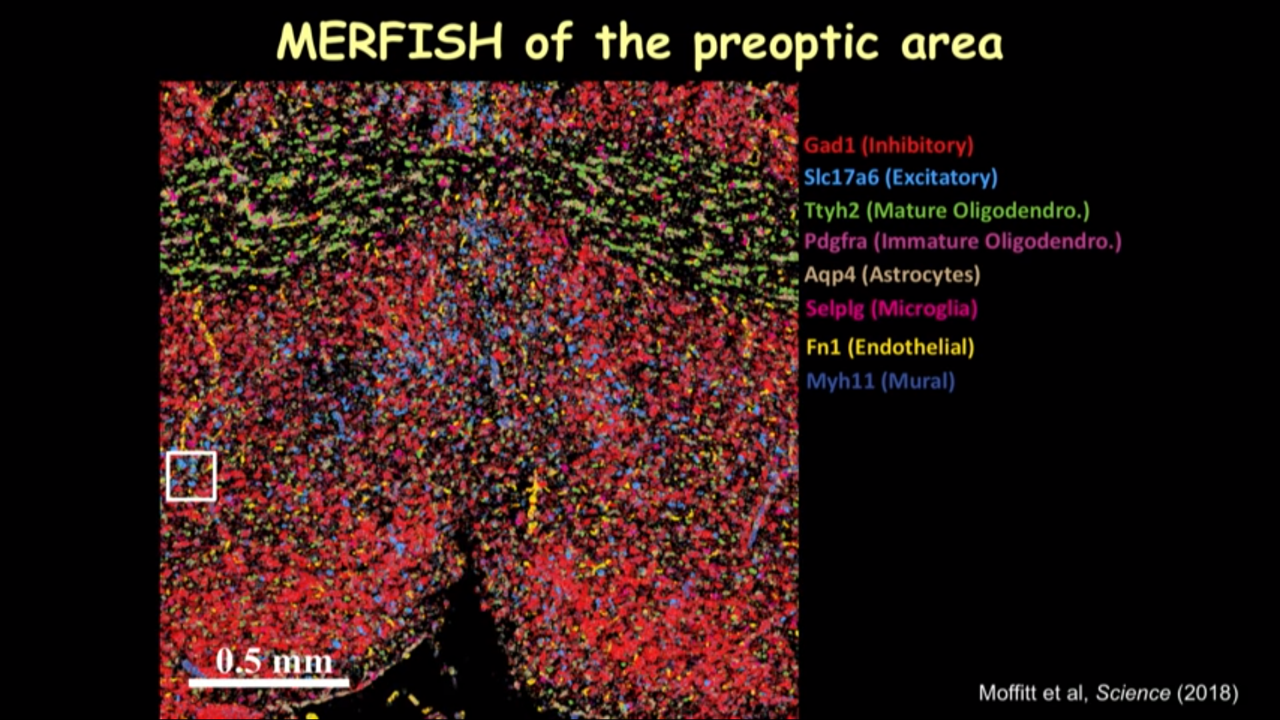
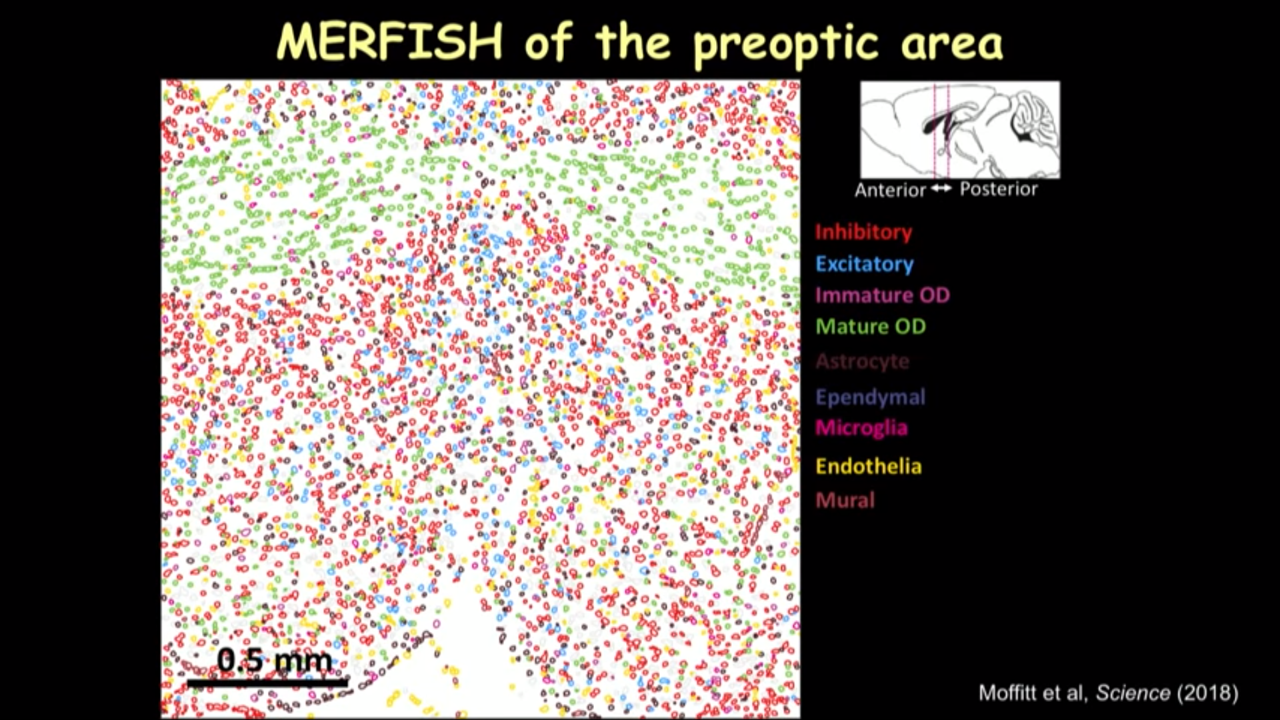
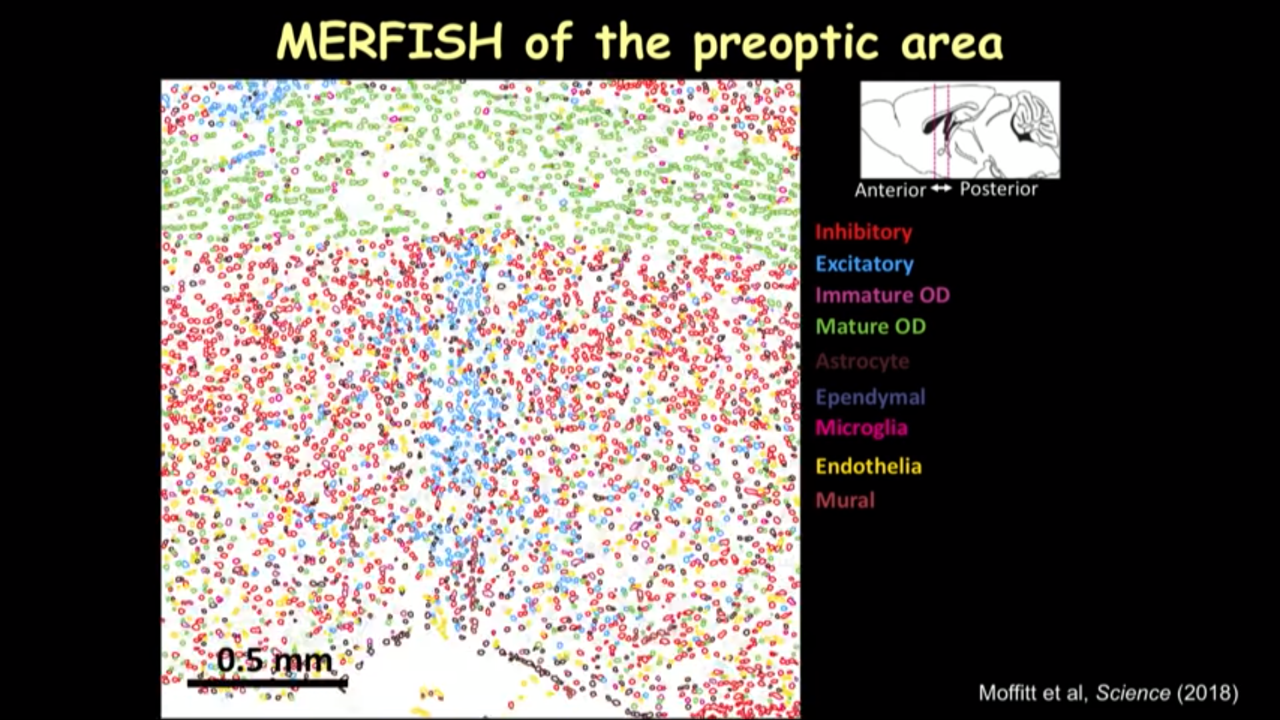
- How MERFISH can help on that front: MERFISH of the preoptic area
- beacuse it is the imaging approach that gets in situ information, they can combine with activity imaging to show the function of these cell types
- After turn on the identity of the genes, computer can actually extract that information
- we can just show several canonical markers and then you can see all these cells have this different color marker(in the image below). It really gave you this direct information on the cell type directly inside the tissue.
- in the imaged much larger region and identified different kinds of cells, when going to move this reigon from the interior to the posterior of the preoptic region to show how dramatically the spatial organization of these cells can change into different locations.
- How MERFISH can help on that front: MERFISH of the preoptic area
- Spatial atlases of cells in complex tissues
 |  |  |
- Application of MERFISH
- Spatial atlases of cells in complex tissues
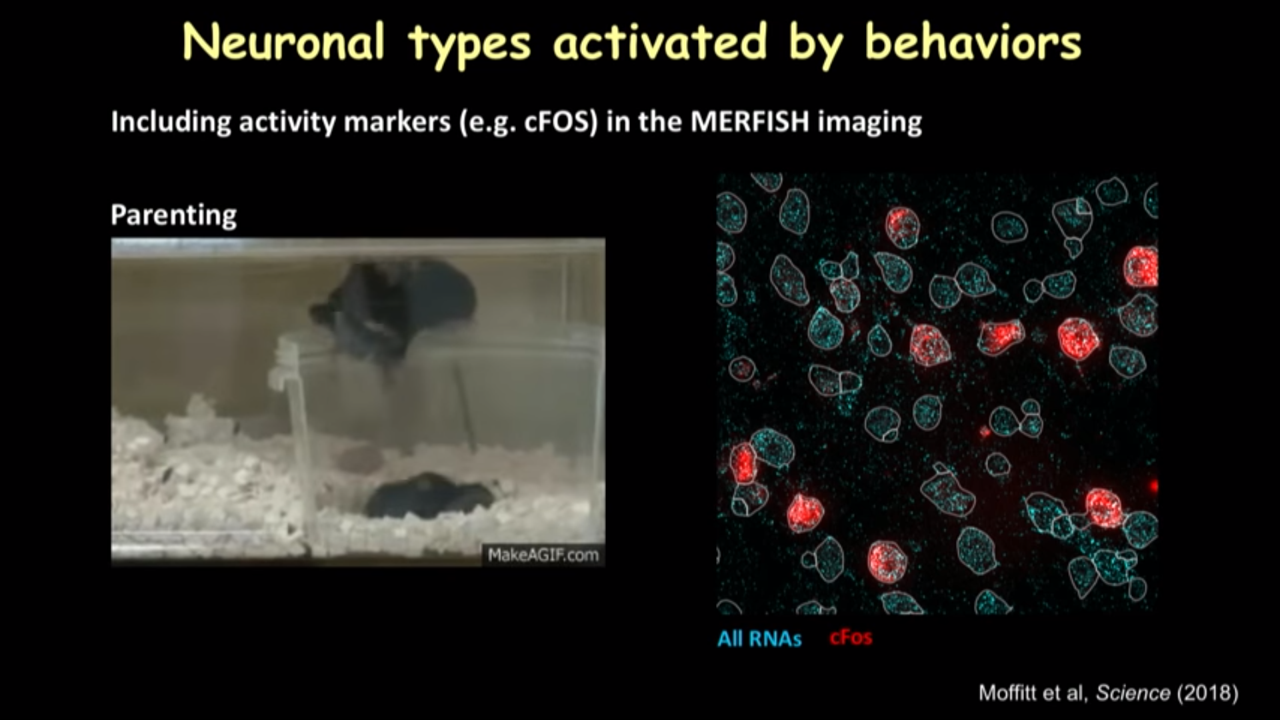
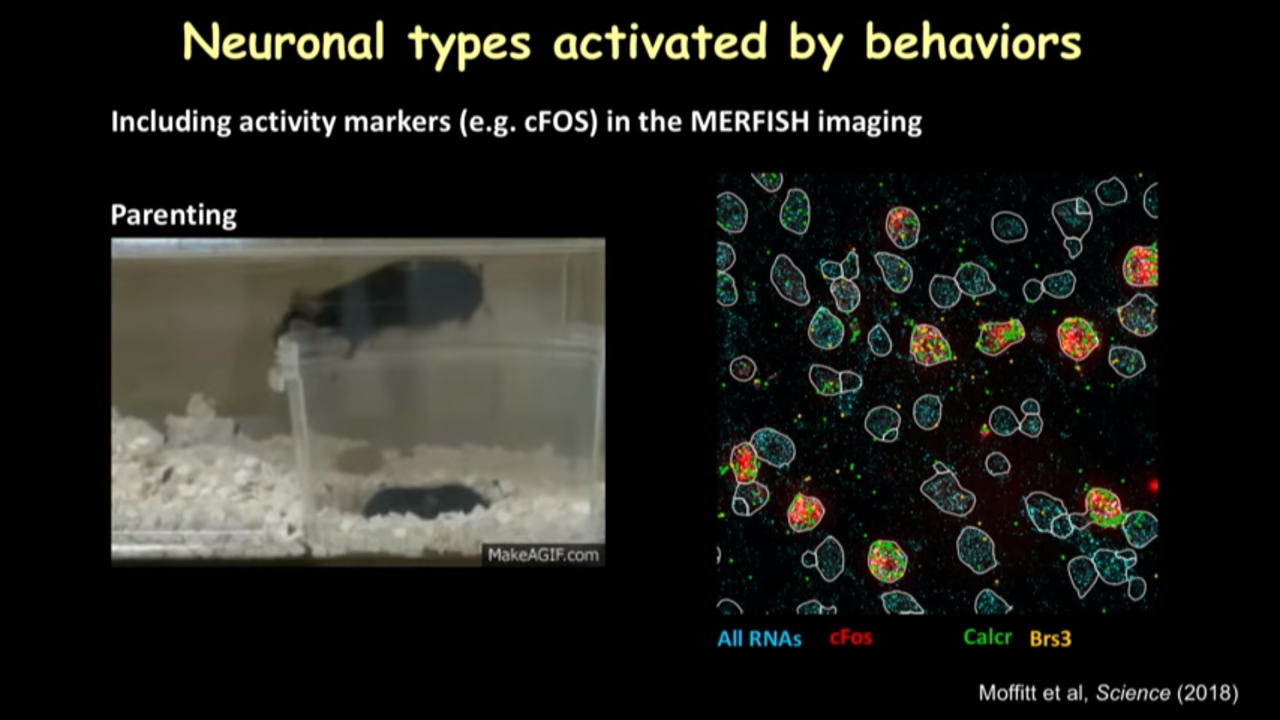
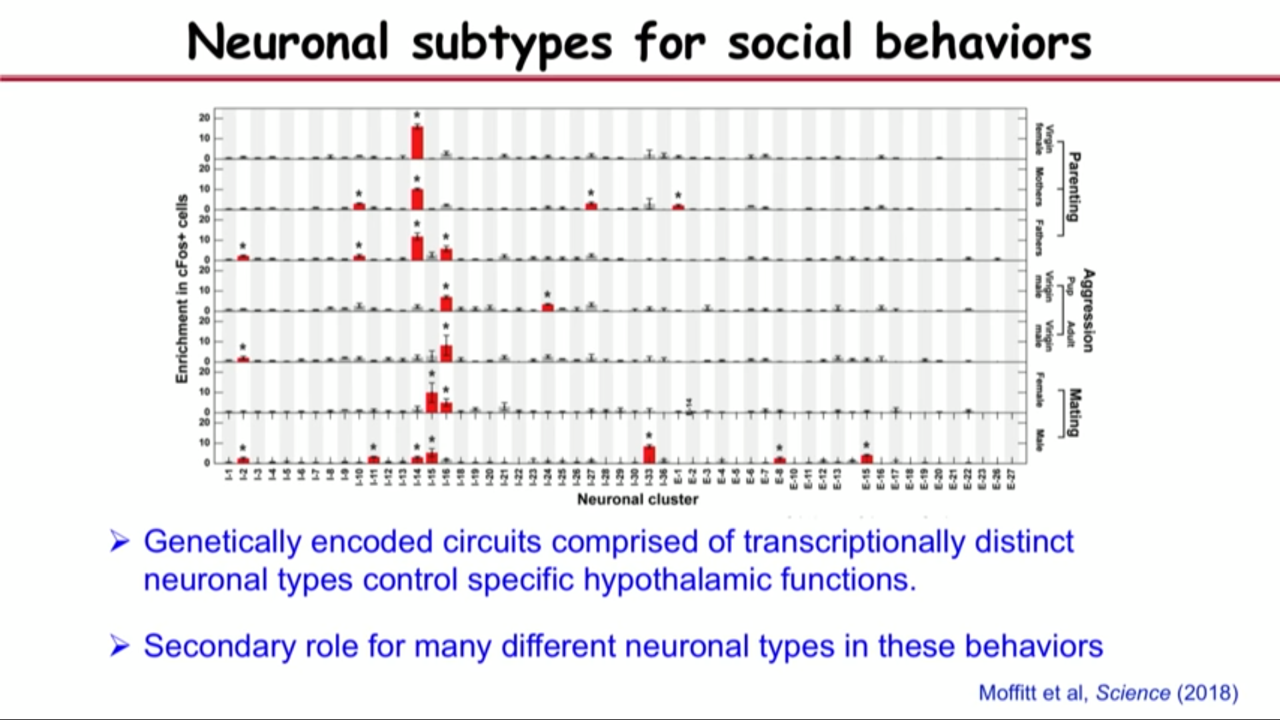
- How MERFISH can help on that front: Neuronal type activated by behaviors
- when the parenting behavior is shown, it actuallu will turn on a kind of a gene called cFOS in the neuronal types that are activated. They include that gene in MERFISH imaging and see the genes are avtivated after parenting.
- they have applied that to study parenting, mating and aggression and identify the neurons in this brain region that are important for the mentioned behavior.
- How MERFISH can help on that front: Neuronal type activated by behaviors
- Spatial atlases of cells in complex tissues
-> A molecular, spatial and functional cell atlas of a brain region
And again, I want to say that there are different approaches to address, different kind of complementary approaches to address this kind of problem.
- e.g. in situ sequenceing
Future challenge: Whole human cell atlas
- how many different types of cells are present
- how are these cells organized
- what are their functions
- how to they communicate with each other
- how do they go wrong in disease
 |
It is their persistence, they are not sort of discouraged by momentary setbacks, their hard work, their strong motivation, their creativity, and that actually mode these kinds of results possible.
The scientific world is getting increasingly collaborative and multidisciplinary. You really need people with complementary expertise to actually join force together to solve problems. And I also listed my remarkable collaborators here. And it’s a very rewarding process.
Others
- special thanks to Ron (Her PhD advisors)
- special thanks to Berkeley (mentors, memories, advisors, friendships, postdoc advisor)
- share some stories with Ron (write papers, teaching, clean manuscript, taught how to write scitific papers)