Get Info from mmCIF
1
2
3
4
5
6
7
8
9
assg_cols = ('_pdbx_struct_assembly_gen.asym_id_list',
'_pdbx_struct_assembly_gen.oper_expression',
'_pdbx_struct_assembly_gen.assembly_id')
oper_cols = ('_pdbx_struct_oper_list.id',
'_pdbx_struct_oper_list.symmetry_operation')
with Path("demo.cif").open('rt') as handle:
mmcif_dict = MMCIF2DictPlus(handle, assg_cols+oper_cols)
Output
2Q4N
1
2
3
4
5
6
{"_pdbx_struct_assembly_gen.assembly_id": ["1"],
"_pdbx_struct_assembly_gen.asym_id_list": ["A,B"],
"_pdbx_struct_assembly_gen.oper_expression": ["1,2"],
"_pdbx_struct_oper_list.id": ["1", "2"],
"_pdbx_struct_oper_list.symmetry_operation": ["x,y,z", "-x,-y+2,z"],
"data_": "2Q4N"}
| Asymmetric unit of 2q4n | Biological assembly 1 of 2q4n |
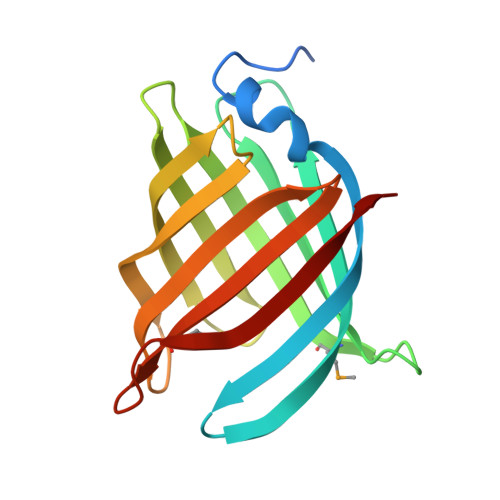 | 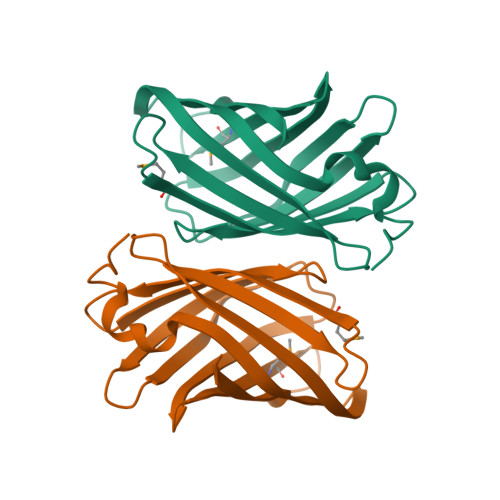 |
3HL2
1
2
3
4
5
6
7
8
9
10
11
{"_pdbx_struct_assembly_gen.assembly_id": ["1", "1", "2", "2"],
"_pdbx_struct_assembly_gen.asym_id_list": ["A,B,F,G,H,I,J,K,Q,R",
"A,B,E,F,G,H,I,J,K,Q,R,U",
"C,D,L,M,N,O,P,S,T",
"C,D,E,L,M,N,O,P,S,T,U"],
"_pdbx_struct_assembly_gen.oper_expression": ["1", "2", "3", "2"],
"_pdbx_struct_oper_list.id": ["1", "2", "3"],
"_pdbx_struct_oper_list.symmetry_operation": ["x,x-y-1,-z",
"x,y,z",
"-x+y,y,-z+1/3"],
"data_": "3HL2"}
| Asymmetric unit of 3hl2 | Biological assembly 1 of 3hl2 | Biological assembly 2 of 3hl2 |
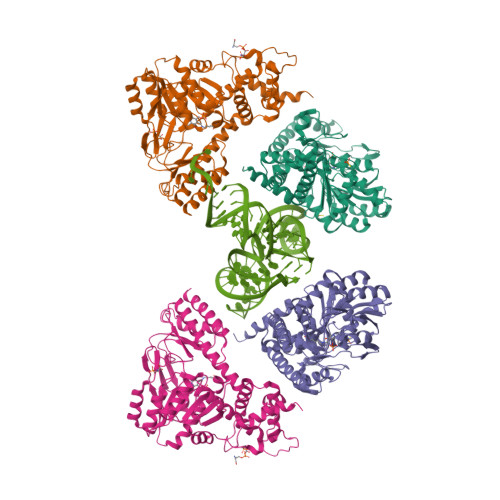 | 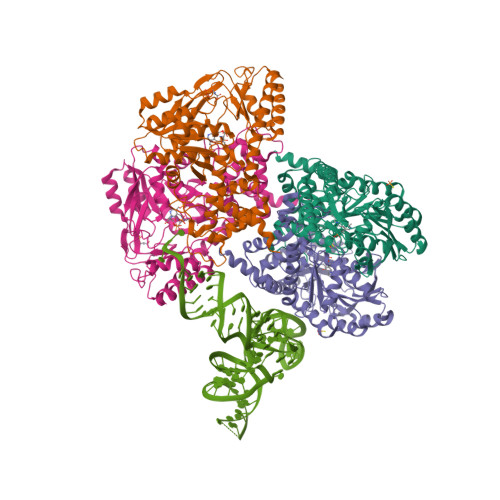 | 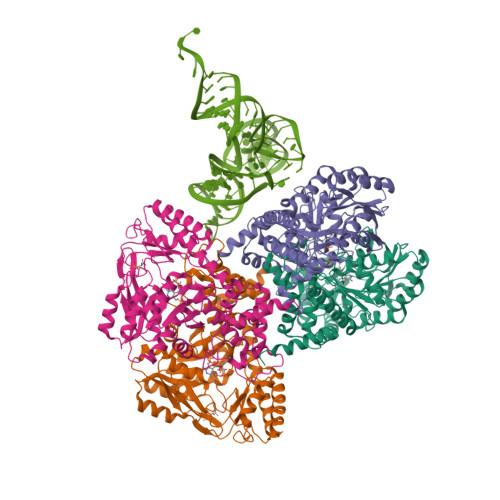 |
1M11
1
2
3
4
5
6
7
8
{
"data_": "1M11",
"_pdbx_struct_assembly_gen.asym_id_list": ["A,B,C,D", "A,B,C,D", "A,B,C,D", "A,B,C,D", "A,B,C,D"],
"_pdbx_struct_assembly_gen.assembly_id": ["1", "2", "3", "4", "5"],
"_pdbx_struct_assembly_gen.oper_expression": ["(1-60)", "1", "(1-5)", "(1,2,6,10,23,24)", "P"],
"_pdbx_struct_oper_list.id": ["P", "1", "2", "3", "4", "5", "6", "7", "8", "9", "10", "11", "12", "13", "14", "15", "16", "17", "18", "19", "20", "21", "22", "23", "24", "25", "26", "27", "28", "29", "30", "31", "32", "33", "34", "35", "36", "37", "38", "39", "40", "41", "42", "43", "44", "45", "46", "47", "48", "49", "50", "51", "52", "53", "54", "55", "56", "57", "58", "59", "60"],
"_pdbx_struct_oper_list.symmetry_operation": ["?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?"]
}
| Asymmetric unit of 1m11 | Biological assembly 1 of 1m11 | Biological assembly 2 of 1m11 |
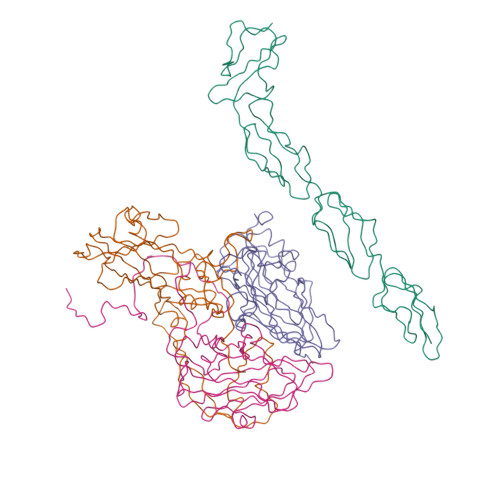 | 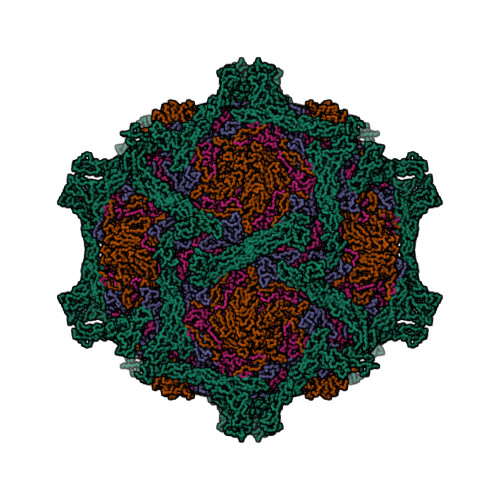 | 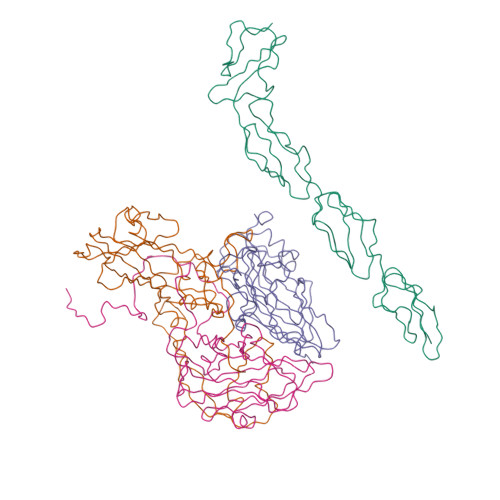 |
| Biological assembly 3 of 1m11 | Biological assembly 4 of 1m11 | Biological assembly 5 of 1m11 |
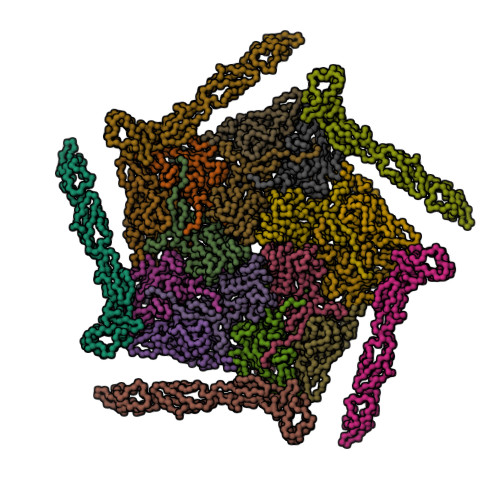 | 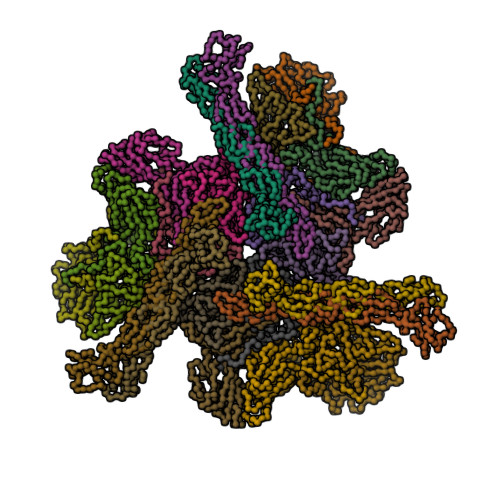 | 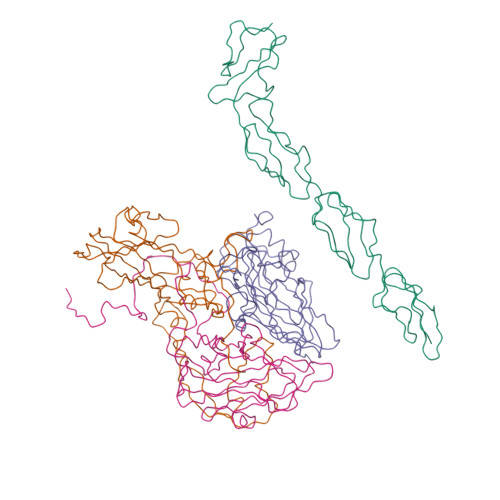 |
1M4X
1
2
3
4
5
6
7
8
{
"data_": "1M4X",
"_pdbx_struct_assembly_gen.asym_id_list": ["A,B,C", "A,B,C", "A,B,C", "A,B,C", "A,B,C", "A,B,C", "A,B,C"],
"_pdbx_struct_assembly_gen.assembly_id": ["1", "2", "3", "4", "5", "6", "7"],
"_pdbx_struct_assembly_gen.oper_expression": ["(1-60)(61-88)", "(61-88)", "(1-5)(61-88)", "(1,2,6,10,23,24)(61-88)", "(1-5)(63-68)", "(1,10,23)(61,62,69-88)", "(P)(61-88)"],
"_pdbx_struct_oper_list.id": ["P", "1", "2", "3", "4", "5", "6", "7", "8", "9", "10", "11", "12", "13", "14", "15", "16", "17", "18", "19", "20", "21", "22", "23", "24", "25", "26", "27", "28", "29", "30", "31", "32", "33", "34", "35", "36", "37", "38", "39", "40", "41", "42", "43", "44", "45", "46", "47", "48", "49", "50", "51", "52", "53", "54", "55", "56", "57", "58", "59", "60", "61", "62", "63", "64", "65", "66", "67", "68", "69", "70", "71", "72", "73", "74", "75", "76", "77", "78", "79", "80", "81", "82", "83", "84", "85", "86", "87", "88"],
"_pdbx_struct_oper_list.symmetry_operation": ["?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?", "?"]
}
1M4X example from https://github.com/biojava/biojava/issues/801#issue-356603172
| Asymmetric unit of 1m4x | Biological assembly 1 of 1m4x | Biological assembly 2 of 1m4x | Biological assembly 3 of 1m4x |
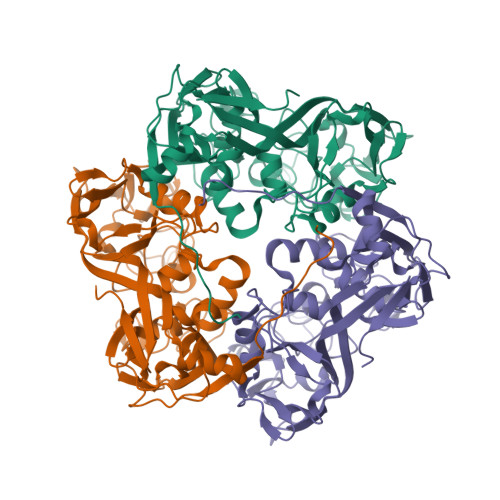 | 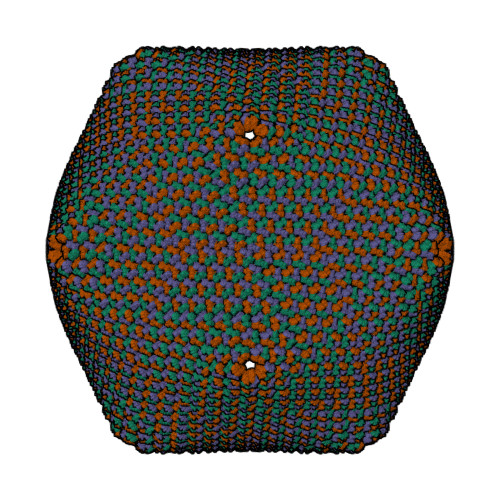 | 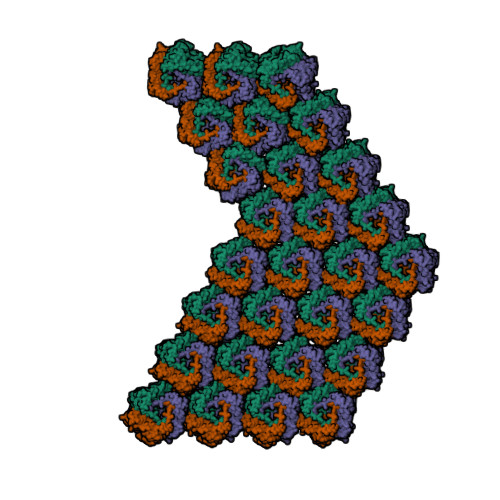 |  |
| Biological assembly 4 of 1m4x | Biological assembly 5 of 1m4x </td> | Biological assembly 6 of 1m4x | Biological assembly 7 of 1m4x |
 |  |  | 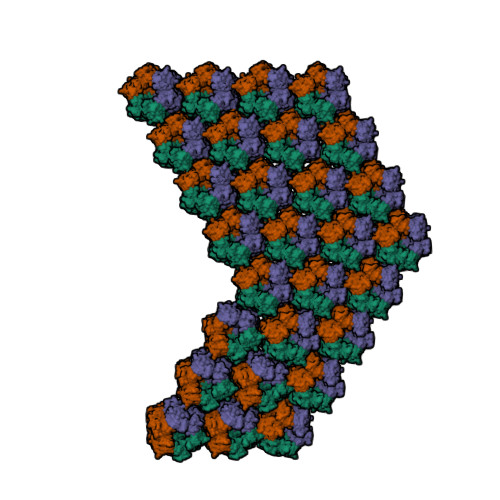 |
How molstar handle assembly info
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
/*
* Code from https://github.com/molstar/molstar/
* src/mol-model-formats/structure/property/assembly.ts
*/
function parseOperatorList(value: string): string[][] {
// '(X0)(1-5)' becomes [['X0'], ['1', '2', '3', '4', '5']]
// kudos to Glen van Ginkel.
const oeRegex = /\(?([^\(\)]+)\)?]*/g, groups: string[] = [], ret: string[][] = [];
let g: any;
while (g = oeRegex.exec(value)) groups[groups.length] = g[1];
groups.forEach(g => {
const group: string[] = [];
g.split(',').forEach(e => {
const dashIndex = e.indexOf('-');
if (dashIndex > 0) {
const from = parseInt(e.substring(0, dashIndex)), to = parseInt(e.substr(dashIndex + 1));
for (let i = from; i <= to; i++) group[group.length] = i.toString();
} else {
group[group.length] = e.trim();
}
});
ret[ret.length] = group;
});
return ret;
}
function expandOperators(operatorList: string[][]) {
const ops: string[][] = [];
const currentOp: string[] = [];
for (let i = 0; i < operatorList.length; i++) currentOp[i] = '';
expandOperators1(operatorList, ops, operatorList.length - 1, currentOp);
return ops;
}
function expandOperators1(operatorNames: string[][], list: string[][], i: number, current: string[]) {
if (i < 0) {
list[list.length] = current.slice(0);
return;
}
let ops = operatorNames[i], len = ops.length;
for (let j = 0; j < len; j++) {
current[i] = ops[j];
expandOperators1(operatorNames, list, i - 1, current);
}
}
//[In]
console.log(expandOperators(parseOperatorList("(X0)(1-4)")));
//[Out]
[ [ "X0", "1" ], [ "X0", "2" ], [ "X0", "3" ], [ "X0", "4" ] ]