ResNetPPI
A lab rotation project. Under development from 2021.12 to 2022.1.
Summary
Predicting protein inter-chain residue distances from sequences irrespective of paired MSA
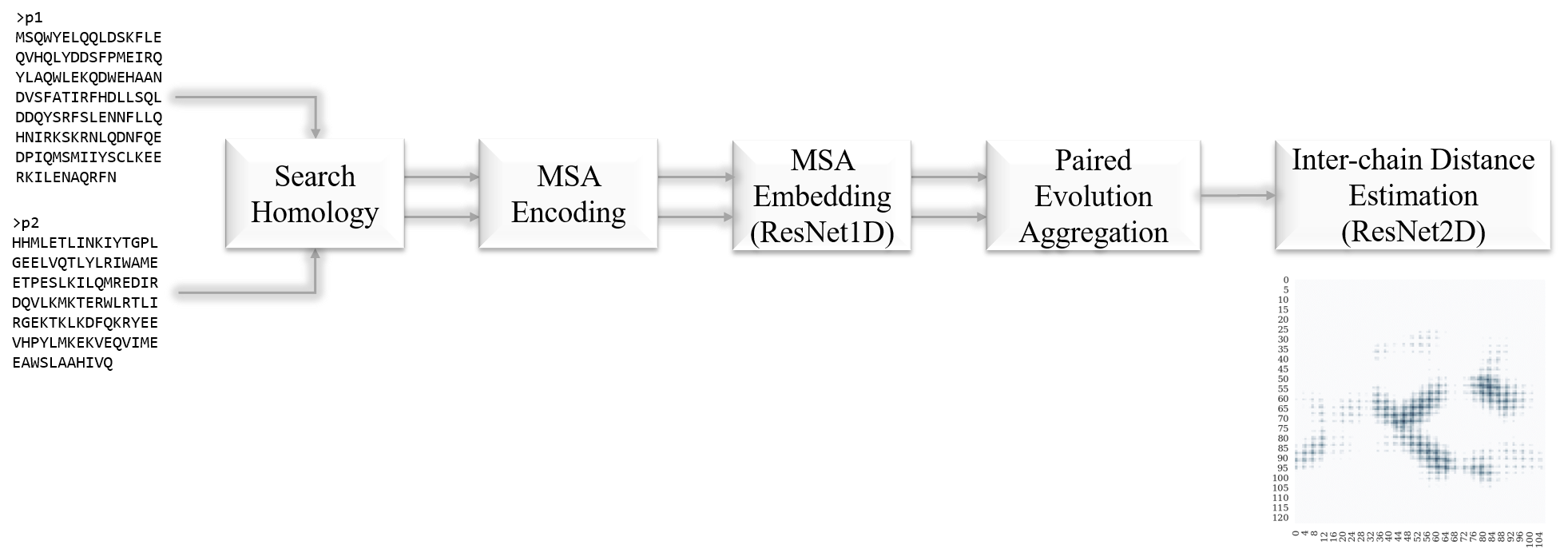
Summary
Advantages
- accept variable input size
- protein of any length
- any number of homologous sequences
- does not rely on paired MSA, thus it can predict cross-species protein interaction
Training
Data Resources
- Sequence database e.g. UniRef30_2020_06_hhsuite.tar.gz
- PDB structures
Input Features
- Protein Sequence * 2
- build MSA via HHblits if possible
- onehot encoding of amino acid type (20+gap+X)*2 + (hydrophoblic+hydrophilic)*2
Fitting Targets
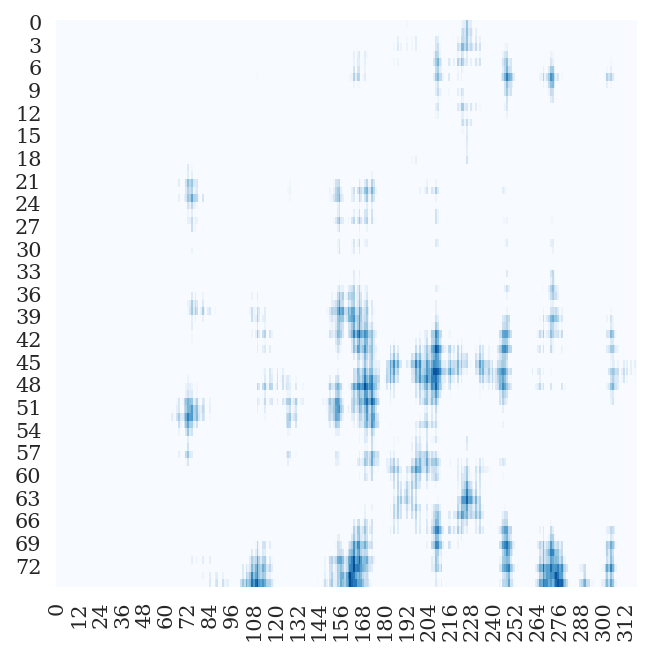
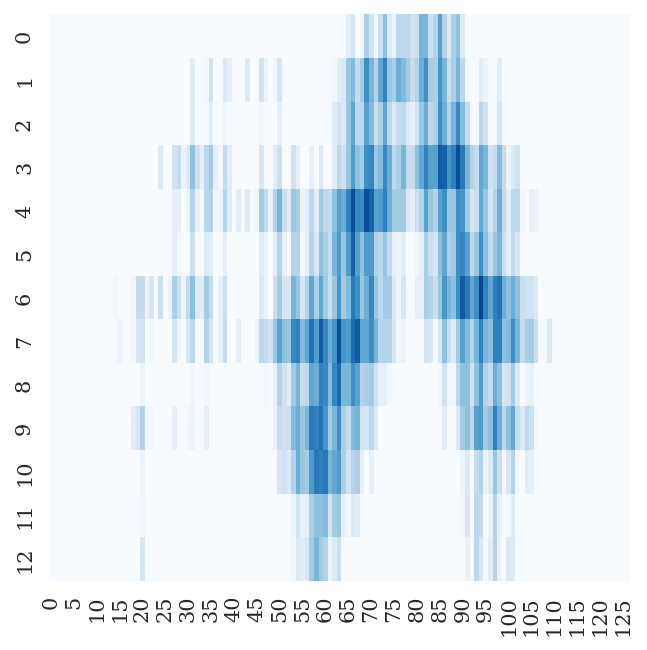
- Fitting Targets (i.e. Inter-chain Cβ-Cβ Distance Map) Examples
- Cα for GLY
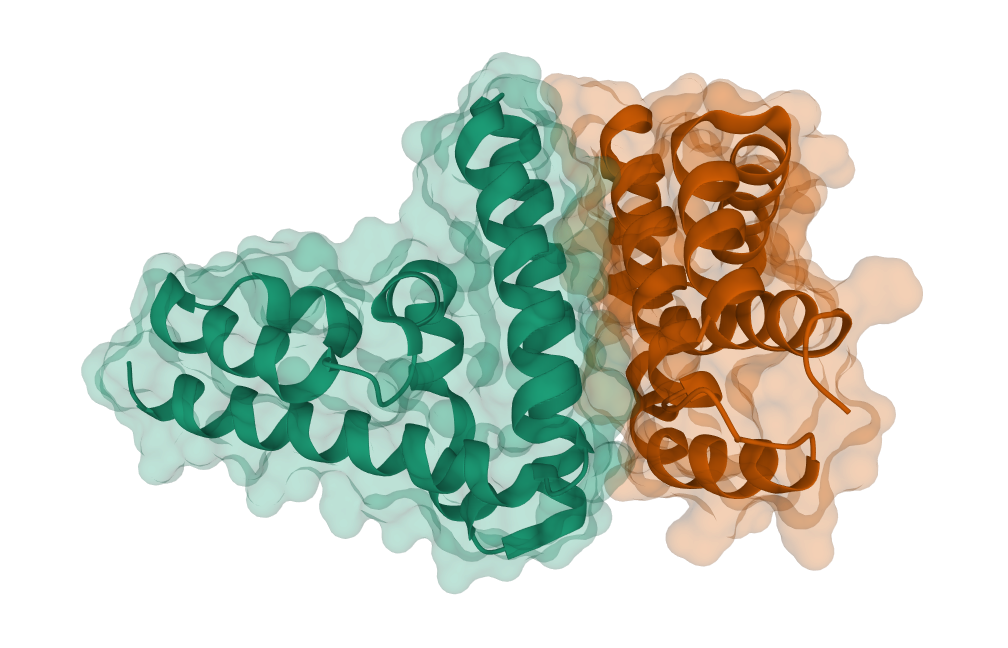
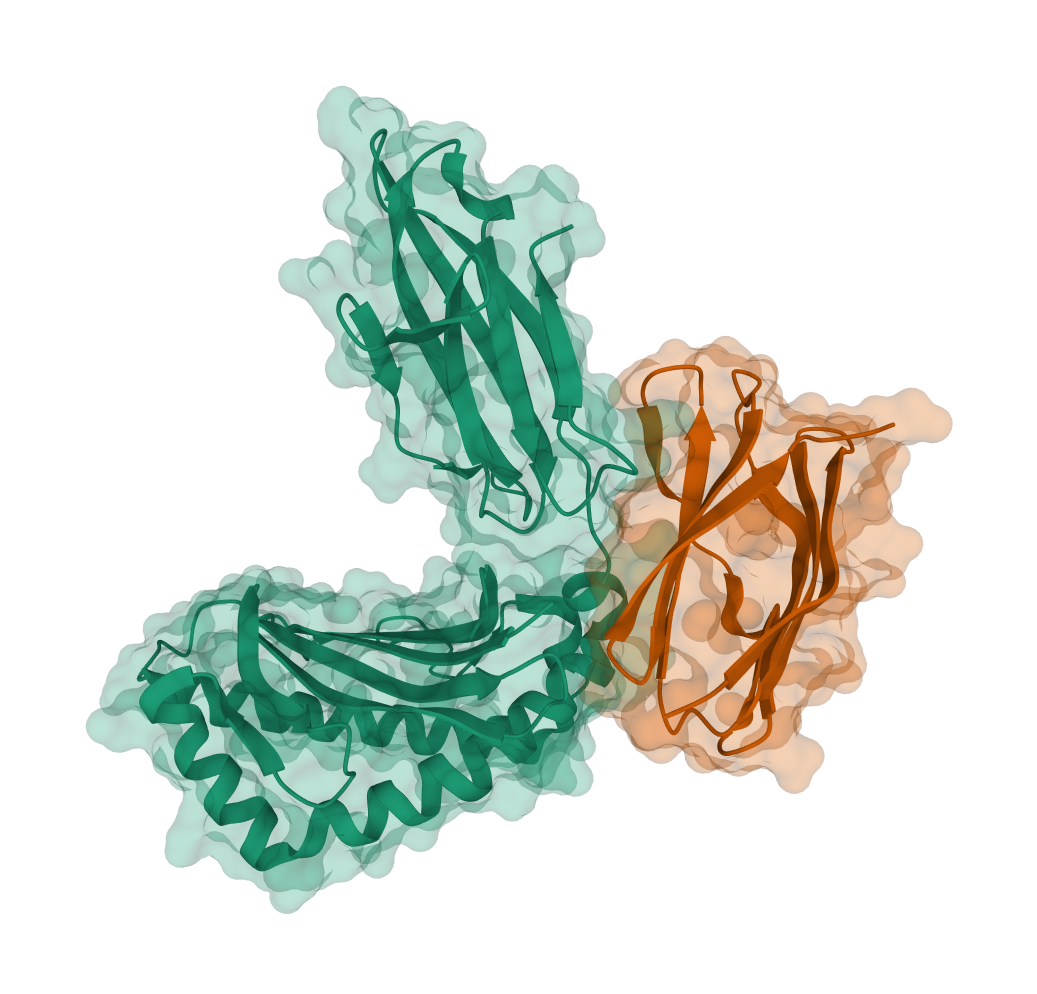
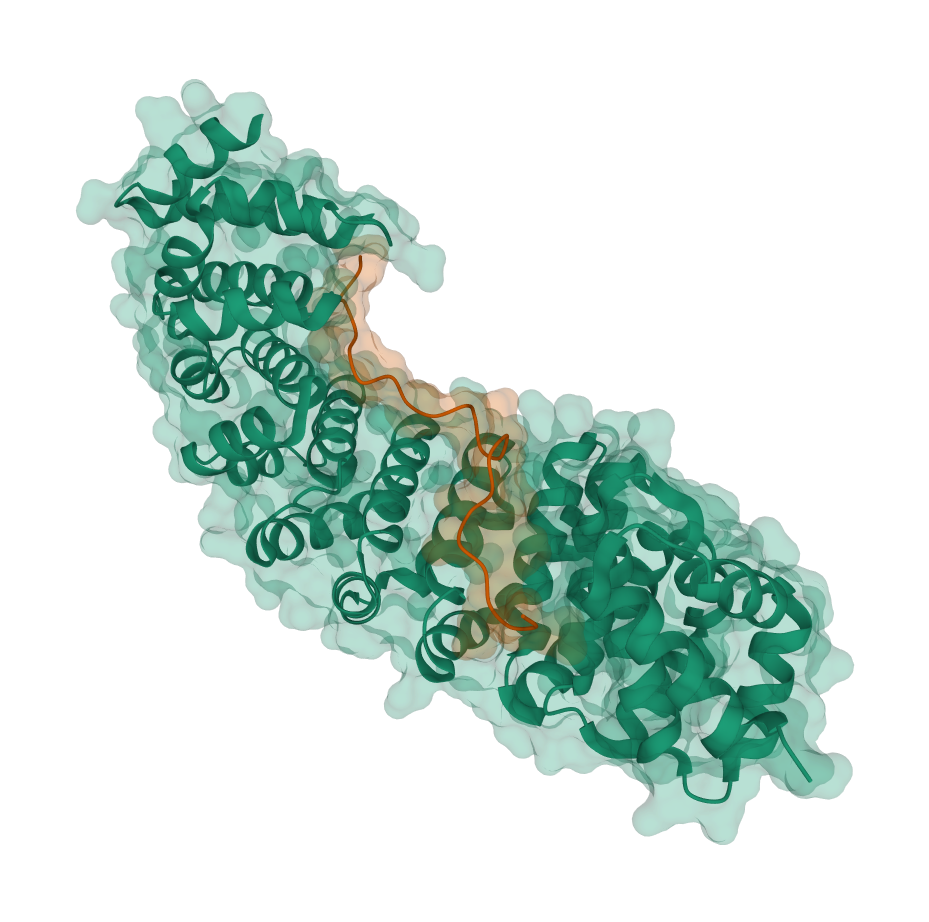
| PDB | Inter-chain Cβ-Cβ Distance Map | pdb_id | human chain | virus chain | len(human chain) | len(virus chain) |
|

|
3wwt | A | B | 123 | 107 |
|

|
1im3 | E | H | 275 | 95 |
|

|
6bvv | A | B | 416 | 24 |

|
|
4rf1 | B | A | 75 | 321 |

|
|
6e5x | B | A | 13 | 127 |
Real-valued distances are discretely binned:
- from 2Å to 20Å
- with bin size of 0.5Å
- 36 bins + 1 bin for \([20, +\infty)\)
Network Architecture
- ResNet1D
- 1d residual block: ((Conv1d + BatchNorm1d) + ELU + (Conv1d + BatchNorm1d)) (+) ELU
- 8 1d-blocks (64 channels)
- ResNet2D
- 2d residual block: ((Conv2d + BatchNorm2d) + ELU + (Conv2d + BatchNorm2d)) (+) ELU
- 16 2d-blocks (96 channels), cycling through dilations \((1,2,4,8)\)
- (mini-)batch size: 1
- Cross-entropy Loss
Model Design
- for each protein sequence (of length \(L\)) search homologous sequences and input the MSA (of size \(K\)) if possible, otherwise input the single sequence
- use original MSA to calculate the weight \(w_k\) for each homologous sequence, \(w_k=\frac{1}{\text{count}(\text{sequence with identity}\ge 0.8)}\)
- calculate \(M_\text{eff}=\sum_{k}^{K}w_k\)
-
MSA Encoding: perform onehot-encoding for each pairwise alignment (\(2\times L_k\), consider both insertion and deletion: \(\rightarrow 48\times L_k\))
- onehot-encoding including 22+2 channels for the reference sequence, 22+2 channels for the homologous sequence
- 22: 20 amino acid types + 1 gap + 1 unknown type
- 2: 1 hydrophoblic + 1 hydrophilic
- for the single sequence input, all the homologous related channels are filled with the reference sequence’s corresponding values
- hence we get \(\{48\times L_k, k\in K\}\)
-
MSA Embedding: for each encoded pairwise alignment, feed into the
ResNet1Dand get embedded pairwise alignment (\(64\times L_k\))- hence we get \(\{64\times L_k, k\in K\}\)
- omit the insertion region of the homologous sequences, thus we can get a \(K\times 64 \times L\) tensor
- thus we have \(x_k\in R^{64\times L}\) and \(x_k(i) \in R^{64}\)
-
Paired Evolution Aggregation
- calculate one body term
- one body term of one sequence: \(f_1(i)=\frac{1}{M_{\text{eff}_{1}}}\sum_{k}^{K_1}w_{1_k} x_{1_k}(i)\)
- one body term of the other sequence: \(f_2(j)=\frac{1}{M_{\text{eff}_{2}}}\sum_{k}^{K_2}w_{2_k} x_{2_k}(j)\)
- apply max function
- \(A(i,c) = \max\{x_k(i,c), k \in K\}\), c: channel; \(A \in R^{64\times L}\)
- calculate two body term
- \[s(i,j) = \frac{1}{\sqrt{M_{\text{eff}_{1}}\cdot M_{\text{eff}_{2}}}}[A_1(i)\otimes A_2(j)], s(i,j)\in R^{64\times 64}\]
- concatenate
- \[h(i,j) = \text{concat}(f_1(i), f_2(j), s(i,j)), h(i,j)\in R^{4224}\]
- calculate one body term
-
Inter-chain Distance Estimation
- feed the \(R^{4224 \times L_1\times L_2}\) tensor into the
ResNet2D - convert the
ResNet2Doutputs into the discrete distance distribution \(37 \times L_1 \times L_2\) throught a (Conv2d+BatchNorm2d+ELU) layer
- feed the \(R^{4224 \times L_1\times L_2}\) tensor into the
During Training
- randomly sample 1000 homologous sequences if \(K > 1000\)
- randomly crop the distance matrix into \(64 \times 64\) shape
- for those input sequences of length \(\le 64\), keep their original length
- loss function ignore those missing residues of PDB structure
- definition of missing
- complete without any modeled atoms for a residue
- GLY without Cα atom
- non-GLY without Cβ atom and without any anchoring atoms (Cα, N, C) that can infer Cβ atom
- definition of missing
Current Training and Validation Results

loss per epoch
Dataset Preparation
- training dataset (362)
- validation dataset (100)
NOTE: the datasets have not been carefully curated, they just serve as demo inputs for training and validation. Thus the training and validation results are just a demonstration of the model’s learning ability.
TODO
- increase mini-batch size
- multiple GPU training
- improve archi
Hardware & Software Prerequisites
- Hardware (tested)
- GPU: NVIDIA Tesla T4 (16G)
- CUDA Version: 11.2
- Software
- Python 3.8 or later in a conda environment
- PyTorch and other Python packages: see requirements
Acknowledgment
I would like to thank Jinyuan Guo and Prof. Huaiqiu Zhu for their helpful discussions and provide me with devices to develop this project.